Project Information and Reproducibility Guide
A pathway for amygdala TMS neuromodulation
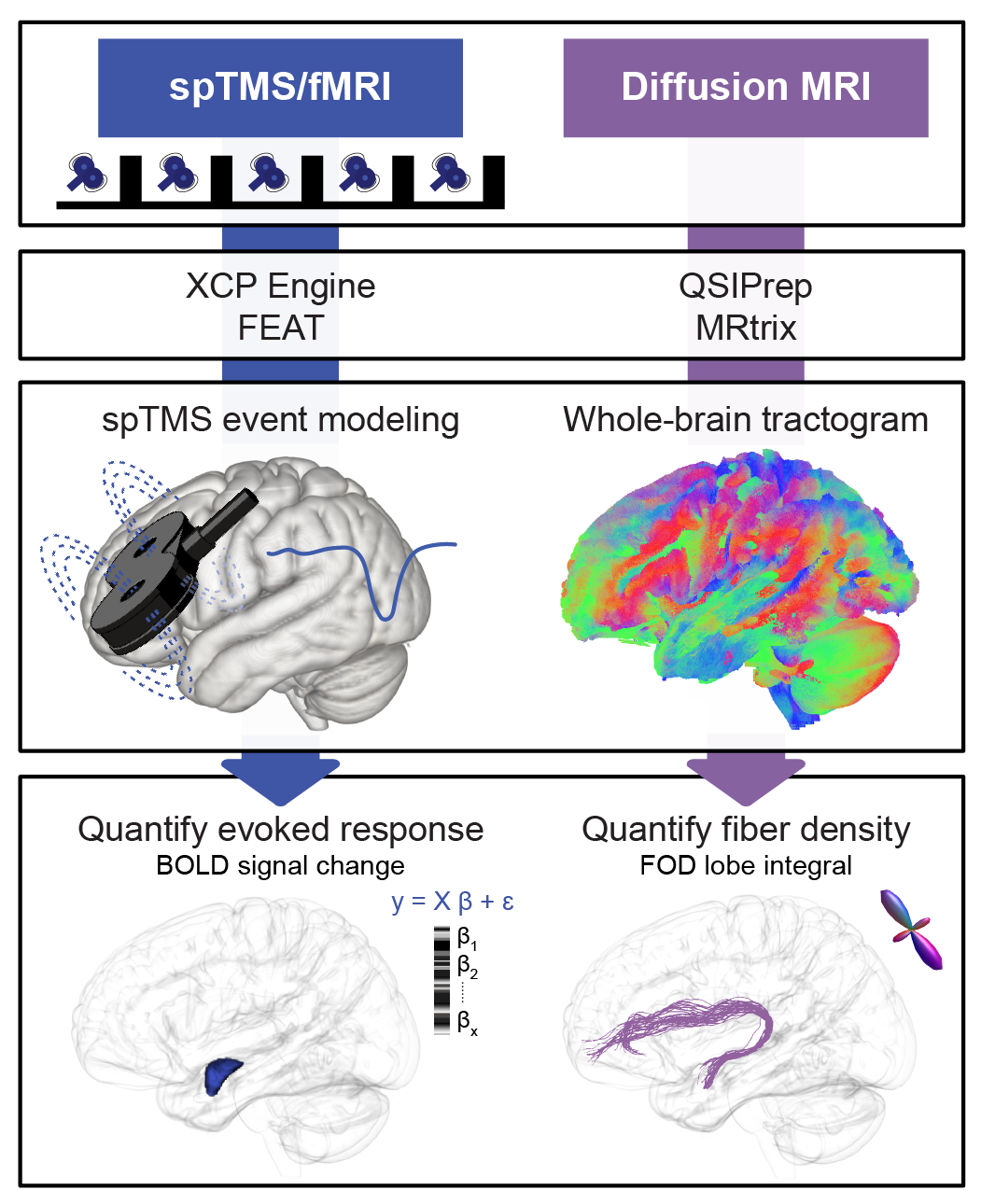
This study demonstrated that transcranial magnetic stimulation applied to the ventrolateral prefrontal cortex (vlPFC) exerts acute and focal neuromodulatory effects on the amygdala by engaging a vlPFC-amygdala white matter pathway.
Project Lead
Valerie J. Sydnor
Faculty Leads
Desmond J. Oathes
Theodore D. Satterthwaite
Analytic Replicator
Matthew Cieslak
Collaborators
Romain Duprat, Hannah Long, Matthew W. Flounders, Joseph Deluisi, Morgan Scully, Nicholas L. Balderston, Yvette I. Sheline, Dani S. Bassett
Project Start Date
August 2018
Current Project Status
Manuscript under Revision
Datasets
ZAPR01-Healthy Controls
Github Repository
https://github.com/PennLINC/ZAPR01_dMRI_TMSfMRI
Path to Data on Filesystem Dopamine
/storage/vsydnor/ZAPR01_WhiteMatter_TMSfMRI
$ZAP/Quality_Control : Study QC spreadsheet
$ZAP/qsiprep_0.6.3 : QSIPrep output
$ZAP/mrtrix_fixelanalysis : output of all mrtrix-based analyses $ZAP/sites_of_stim : TMS SOS coordinates, ROIs, masks
$ZAP/templates : templates and ROI masks
$ZAP/output_measures : spreadsheets with study measures for stats
Conference Presentations
Poster presentation at The Society of Biological Psychiatry Annual Meeting, April 2021. Amygdala TMS-fMRI Evoked Response is Influenced by Prefrontal-Amygdala White Matter Pathway Fiber Density
CODE DOCUMENTATION
The analytic workflow implemented in this project is described in detail in the following sections. Analysis steps are described in the order they were implemented; the script(s) used for each step are identified and links to the code on github are provided.
Diffusion MRI Preprocessing and Reconstruction
ZAPR01 diffusion MRI data was first BIDSifyed, preprocessed with QSIPrep, and postprocessed with mrtrix’s fixel-based analysis pipeline, as detailed below:
- Organize Data into BIDS
- ZAPR01 data was organized into the Brain Imaging Data Structure via heudiconv-0.5.4 via the script /bids/ZAP2BIDS.sh. This script both converts the original ZAPR01 directory/file structure to BIDS and adds IntendedFor specifications to fieldmap jsons
- The study-specific heuristic /bids/ZAPR01_legacyandnew_heuristic.py was used for BIDS conversion via the call
$ sh ZAP2BIDS.sh vsydnor /data/jux/oathes_group/projects/vsydnor/scripts/ZAP2BIDS/ZAPR01_legacyandnew_heuristic.py - Note: heudiconv was run on chead (BIDS output on chead: /data/jux/oathes_group/studies/TNI_ZAPR01/MRI/BIDS)
- Preprocess Diffusion Data with QSIPrep
- ZAPR01 single shell diffusion data were preprocessed with qsiprep version 0.6.3RC3
- First, the singularity image qsiprep-0.6.3RC3.simg was built from docker by executing /qsiprep/build_qsiprep_0.6.3.simg.sh
- QSIPrep was then run via a job array by executing /qsiprep/run_qsiprep0.6.3_ZAPR01_DTI.sh. QSIPrep was run with the following parameters:
$ qsiprep-0.6.3RC3.simg --bids_dir $bidsdir --output_dir $qsiprepdir --analysis_level participant --participant_label $SUBJ --hmc_model eddy --eddy-config eddy_params.json --b0-motion-corr-to first --output-space T1w --output-resolution 1.3 --force-spatial-normalization --do-reconall - Note: qsiprep-0.6.3RC3.simg was built and run on chead, and the output of qsiprep was copied to dopamine (/storage/vsydnor/ZAPR01_WhiteMatter_TMSfMRI/qsiprep_0.6.3). All subsequent scripts/analyses were run on dopamine
- Postprocess Diffusion Data with MrTrix Fixel-Based Analysis Pipeline
The mrtrix fixel-based analysis pipeline was implemented to postprocess the output of qsiprep. This pipeline includes diffusion signal reconstruction via constrained spherical deconvolution, fixel segmentation, and tractography generation. The pipeline generates subject-specific FOD images and a study-specific FOD template, subject-specific fixel images and a study-specific fixel template, and template-based whole-brain tractography. The pipeline outlined below includes the relevant steps documented in mrtrix3Tissue and mrtrix CSD fixel pipeline and was executed as follows:
- Diffusion images preprocessed with QSIPrep were converted from nifti to mif by /mrtrix_csd_fixels/mrconvert_nii2mif_1.sh
- Diffusion images underwent initial bias field correction via /mrtrix_csd_fixels/biasfield_correction_2.sh to enhance the quality of brain masking
- Diffusion image brain masks were generated with /mrtrix_csd_fixels/dwi2mask_3.sh
- WM, GM, and CSF response functions were computed for each subject by /mrtrix_csd_fixels/responsefunction_dhollander_4.sh, which calls mrtrix3Tissue’s dwi2response single-shell 3-tissue response function estimation algorithm
- A unique set of study-specific WM, GM, and CSF response functions were generated by averaging response functions from all study subjects via /mrtrix_csd_fixels/average_responsefunction_5.sh
- Subject-specific FOD images (and GM and WM images) were reconstructed via 3-tissue constrained spherical deconvolution modeling (ss3t_csd_beta1) by running /mrtrix_csd_fixels/ss3t_csd_6.sh
- FOD images (and GM and WM images) underwent bias field correction and global intensity normalization with mtnormalise via /mrtrix_csd_fixels/mtnormalise_7.sh
- A study-specific FOD template was computed by executing /mrtrix_csd_fixels/FOD_populationtemplate_8a.sh (set up directory structure) and /mrtrix_csd_fixels/FOD_populationtemplate_8b.sh (run population_template), using data from all 45 study subjects
- Warps from subject space to the study-specific FOD template were computed with /mrtrix_csd_fixels/subjectFOD_to_ZAPR01FODTemplate_warp_9.sh
- An FOD template mask was generated by running /mrtrix_csd_fixels/FOD_populationtemplate_mask_10.sh
- A study-specific Fixel template was created by segmentating template FODs with /mrtrix_csd_fixels/FixelMask_populationtemplate_11.sh
- Subject-specific FOD images were registered from subject space to the study-specific FOD template by applying pre-computed warps in /mrtrix_csd_fixels/subjectFOD_to_ZAPR01FODTemplate_applywarp_12.sh
- Subject-specific fixel masks were generated in template space, FD was computed, and fixels were reoriented to template fixels via /mrtrix_csd_fixels/subjectFixels_FD_13.sh
- Subject fixels were assigned to study-specific template fixels with /mrtrix_csd_fixels/fixelcorrespondence_14.sh
- FC and FDC were computed for subject fixels with /mrtrix_csd_fixels/subjectFixels_FC_FDC_15.sh
- Whole-brain tractography was performed with /mrtrix_csd_fixels/Tractography_populationtemplate_16.sh using the study-specific FOD template as input and parameters recommended in the mrtrix documentation
- A fixel-fixel connectivity matrix was generated, based on the whole-brain tractography, to enable fixel measure smoothing via /mrtrix_csd_fixels/fixelconnectivity_17.sh
- Fixel-based measures were smoothed with /mrtrix_csd_fixels/fixelconnectivity/fixelsmoothing_18.sh
TMS Sites of Stimulation – Left Amygdala Tract Extraction
Following preprocessing, FOD/fixel reconstruction, and tractography, the whole-brain tractography was used to delineate a putative causal pathway by which TMS-evoked cortical activity could travel to the left amygdala. To identify this pathway, a study-specific TMS cortical sites of stimulation mask was generated, and streamlines with endpoints in this mask and in the left amygdala were extracted, as follows:
- Generate a Cortical Sites of Stimulation Mask for Amygdala-targeted TMS
- For each subject, the TMS site of stimulation for amygdala-targeted TMS was localized to a set of X, Y, Z MNI space voxel coordinates using Brainsight NeuroNavigation information. A site of stimulation spherical ROI was generated from the MNI coordinates, and ROIs from all subjects were merged into a single TMS sites of stimulation mask. This was accomplished with the script /sites_of_stim/siteofstim_processing.sh
- Register Tractography Inclusion Masks from MNI Space to FOD Template
To identify tractography streamlines with endpoints in the amygdala TMS sites of stimulation mask and the left amygdala, the sites of stimulation mask (generated in step 1) and a left amygdala ROI (extracted from the Harvard Oxford subcortical atlas) were transformed from MNI space to the study-specific FOD template
- This was accomplished by first registering the HCP1065 FA template from MNI space to a study-space FA template in FOD template space with /tract_analyses/MNI_to_ZAPR01FODTemplate_warp.sh, and then applying the transforms to the masks of interest with /tract_analyses/MNImasks_to_ZAPR01FODTemplate_applywarp.sh
- Extract Causal Pathway Streamlines
- Streamlines connecting the amygdala TMS sites of stimulation mask and the left amygdala were extracted with /tract_analyses/tracts_amygdalaSOSmask_LHamygdalaROI_tckedit.sh, identifying a left vlPFC-amygdalar white matter pathway in humans
- Map Pathway Streamlines to Fixels
- In order to quantify mean fixel-based pathway measures for each subject (e.g. fiber density), the causal white matter pathway streamlines were mapped back to template fixels with /tract_analyses/tracts_to_fixels.sh
- Tract fixels were then cropped from the whole-brain fixed mask via /tract_analyses/fixelcrop.sh (using a primary streamline threshold of 5 as well as additional streamline thresholds for sensitivity analyses)
Tract Anatomy and Brodmann Overlap
The scripts in /tract_analyses/anatomy were written to gain insight into the anatomy of the identified vlPFC-amygdala pathway.
- Pathway streamlines were mapped to voxels for the visualization in Figure 3B using tracts_to_voxels.sh
- The Brodmann areas that pathway streamlines originated in were determined by running tractendpoints.sh followed by tractorigins_brodmann.sh
- Overlap in the trajectory of the vlPFC-amygdala pathway and JHU major white matter tracts was calculated with tracttrajectory_JHU.sh
Tract-Based Mean Fixel Measures and TMS-fMRI Evoked Response Measures
To assess whether microstructural (FD) and macrostructural (logFC) properties of the vlPFC-amygdala white matter pathway were associated with the magnitude of TMS-evoked functional response in the amydala (and to conduct related sensitivity and specificity analyses), vlPFC-amygdala white matter pathway fixel measures (FD, logFC, FDC) were extracted for each subject with /tract_analyses/fixelmeasures.sh and TMS-fMRI functional evoked response data were extracted with /TMSfMRI_EvokedResponse/TMSfMRI_SignalChange_TMSon_Measures.sh. Note: the fMRI data were preprocessed with fMRIPrep and postprocessed with XCP to generate single pulse TMS-fMRI BOLD signal change (i.e. evoked response) maps, as detailed in Duprat et al. (In preparation).
Statistical Analysis
Manuscript statistics were conducted in R and are included in /statistics/dMRI_TMSfMRI_statistics.Rmd. All statistics were computed with the data provided in the master study spreadsheet Sydnor2022_ZAPR01_Amygdala_TMSfMRIdMRI.csv
Visualization
Manuscript figures were generated using R, mrview, fsleyes, and Slicer. Slicer was used to visualize TMS sites of stimulation (Figure 2A, Figure 4C); the center of gravity for all vlPFC stimulation sites / control stimulation sites was calculated with /sites_of_stim/stimsites_vlPFC_control_COG.sh. Mrview was used to visualize the vlPFC-amygdala pathway, as well as pathway streamlines, FODs, and fixels (Figure 3A). Fsleyes was used to visualize pathway anatomy overlaid on the JHU white matter atlas (Figure 3B). The R code in /visualization/dMRI_TMSfMRI_visualization.Rmd was written to generate all graphs (Figure 2B, Figure 2C, Figure 4A, Figure 4B, Figure 4D).