Data Narrative for DAY2 - Margaret’s Curation
Data Processing Flow & Important Links:
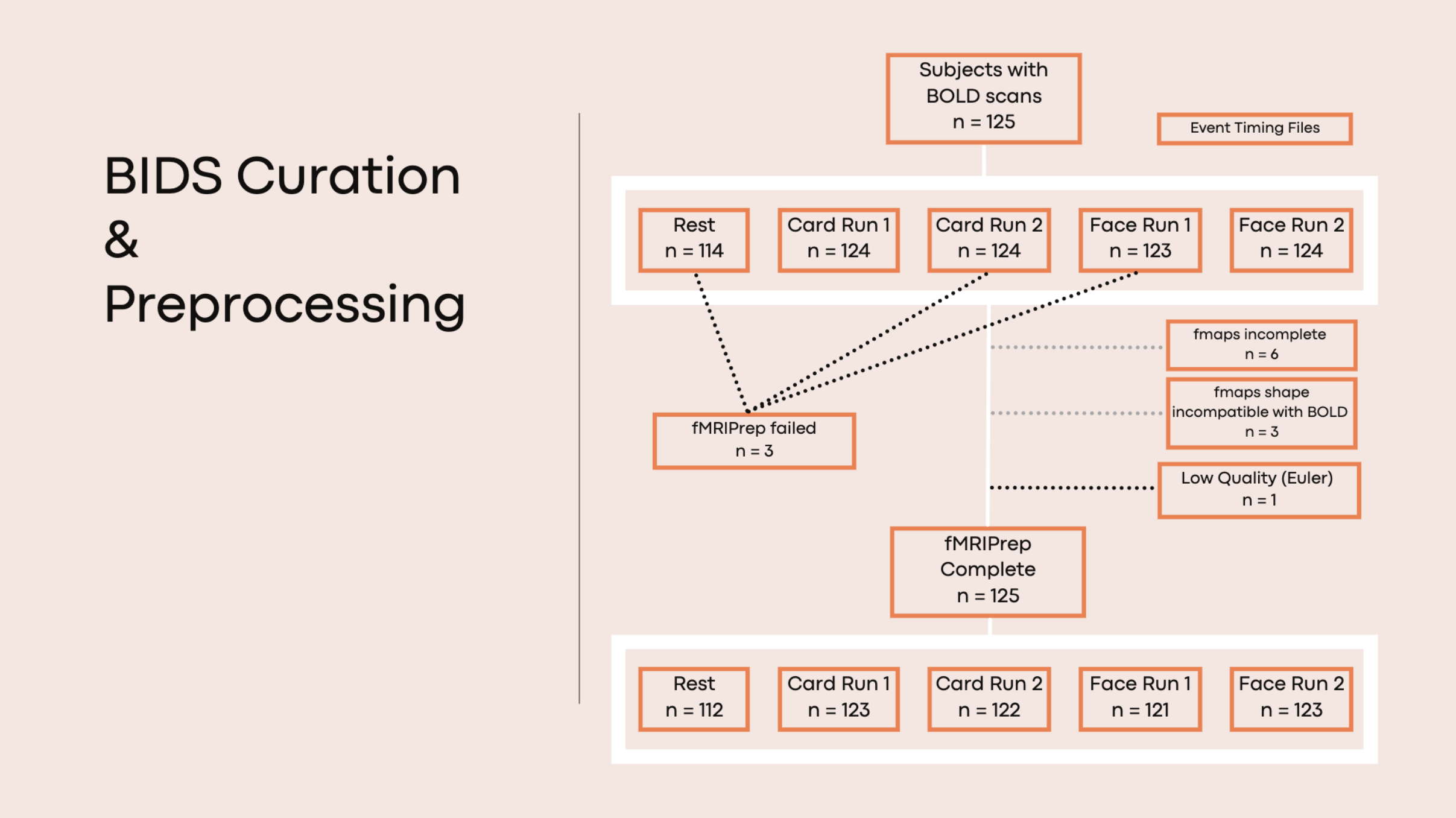
- Flow diagram that describes the lifecycle of this dataset curation and preprocessing may be viewed here
- Overview:
- subjects without usable fmaps (no SDC run in fMRIPrep): ‘sub-12235’ ‘sub-13585’ ‘sub-14610’ ‘sub-14848’ ‘sub-14858’ ‘sub-14876’ ‘sub-15546’ ‘sub-16181’ ‘sub-16234’ ‘sub-17726’ *scans noted here
- subjects/scans that failed fMRIPrep: sub-13373_ses-day2_task-face_run-01_bold.nii.gz, sub-14858_ses-day2_task-card_run-02_bold.nii.gz, sub-15709_ses-day2_task-rest_bold.nii.gz note: gzip error, can be rerun with later version
- subjects/scans with poor QC: sub-15433 T1w (euler=782); sub-17378 card1, card2, face1, face2 (normCrossCorr <0.8); sub-15276 face2 (normCrossCorr <0.8); sub-15433 card 2 (normCrossCorr <0.8); note: paths to XCP-generated .html reports for each subject and concatenated qc values provided below
Plan for the Data
- Why does PennLINC need this data?
- For which project(s) is it intended? Please link to project pages below:
- Goal is to curate and preprocess data
Data Acquisition
- Data acquired by Dan Wolf
- Describe the data:
- number of subjects = 125
- types of images = bold (2 runs task-face, 2 runs task-card, rest), T1w, T2w, DWI note: run1=task version A and run2 = task version B according to json SeriesDescription, see task-match.ipynb
- T1w = 125 subj
- T2w = 3 subj
- card_run-01 = 124 subj
- card_run-02 = 124 subj
- face_run-01 = 123 subj
- face_run-02 = 124 subj
- rest = 114 subj
- fmap = 124 subj
- dwi = 3 subj
Download and Storage
- Data was stored as nifti files in
/cbica/projects/wolf_satterthwaite_reward/original_data/bidsdatasets/day2. - Data was copied by Margaret to sub-project folder
/cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/original_dataon 9/14/2021. JSON’s within origial_data were updated using
cubids-add-nifti-info.- Listing metadata fields using
cubids-print-metadata-fieldsresulted:- Acknowledgements
- AcquisitionMatrixPE
- AcquisitionNumber
- Authors
- BIDSVersion
- BandwidthPerPixelPhaseEncode
- BaseResolution
- CoilString
- ConversionSoftware
- ConversionSoftwareVersion
- DatasetDOI
- DeidentificationMethod
- DerivedVendorReportedEchoSpacing
- DeviceSerialNumber
- Dim1Size
- Dim2Size
- Dim3Size
- DwellTime
- EchoNumber
- EchoTime
- EchoTime1
- EchoTime2
- EchoTrainLength
- EffectiveEchoSpacing
- FlipAngle
- Funding
- HowToAcknowledge
- ImageOrientationPatientDICOM
- ImageType
- ImagingFrequency
- InPlanePhaseEncodingDirectionDICOM
- IntendedFor
- InversionTime
- License
- MRAcquisitionType
- MagneticFieldStrength
- Manufacturer
- ManufacturersModelName
- Modality
- Name
- NumVolumes
- Obliquity
- ParallelReductionFactorInPlane
- PartialFourier
- PercentPhaseFOV
- PhaseEncodingDirection
- PhaseEncodingSteps
- PhaseResolution
- PixelBandwidth
- ProcedureStepDescription
- ProtocolName
- PulseSequenceDetails
- ReceiveCoilName
- ReconMatrixPE
- RefLinesPE
- ReferencesAndLinks
- RepetitionTime
- SAR
- ScanOptions
- ScanningSequence
- SequenceName
- SequenceVariant
- SeriesDescription
- SeriesInstanceUID
- SeriesNumber
- ShimSetting
- SliceThickness
- SliceTiming
- SoftwareVersions
- SpacingBetweenSlices
- TaskName
- TotalReadoutTime
- TxRefAmp
- VoxelSizeDim1
- VoxelSizeDim2
- VoxelSizeDim3
- template
Running
cubids-remove-metadata-fieldsresulted no PHI fields for removal. - Data checked into DataLad
/cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/curation/BIDS (dataset)viadatalad save -m "add initial data" -d ./curation/BIDSaction summary: add (ok: 2448) save (ok: 1)
Curation Process
- Data curation by Margaret Gardner for NGG rotation on the CUBIC project user
wolfsatterthwaitereward - Link to final CuBIDS csvs:
/cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/curation/code/iterations/iteration6
BIDS Validation:
Iteration 1 (Ran
cubids-validateandcubids-groupsimultanously as per The WAY, outputs saved tosandbox/validator_outputs/iteration1): EVENTS_TSV_MISSING ( Task scans should have a corresponding events.tsv file. If this is a resting state scan you can ignore this warning or rename the task to include the word “rest”. ) : 495 counts INCONSISTENT_SUBJECTS ( Not all subjects contain the same files. Each subject should contain the same number of files with the same naming unless some files are known to be missing. ) : 806 counts INCONSISTENT_PARAMETERS ( Not all subjects/sessions/runs have the same scanning parameters. ) : 24 subjects README_FILE_MISSING ( The recommended file /README is missing. See Section 03 (Modality agnostic files) of the BIDS specification. ) : 1 subjects NO_AUTHORS ( The Authors field of dataset_description.json should contain an array of fields - with one author per field. This was triggered because there are no authors, which will make DOI registration from dataset metadata impossible. ) : 1 subjectsIteration 1.2 (Reran
cubids-validatewith--ignore_nifti_headersand--ignore_subject_consistency, no modifications to datafiles): EVENTS_TSV_MISSING ( Task scans should have a corresponding events.tsv file. If this is a resting state scan you can ignore this warning or rename the task to include the word “rest”. ) : 495 scans README_FILE_MISSING ( The recommended file /README is missing. See Section 03 (Modality agnostic files) of the BIDS specification. ) : 1 count NO_AUTHORS ( The Authors field of dataset_description.json should contain an array of fields - with one author per field. This was triggered because there are no authors, which will make DOI registration from dataset metadata impossible. ) : 1 count*counts using validator_err_counts.ipynb
BIDS curation approved by Ted Satterthwaite and Tinashe Tapera on 9/21/21, last validator output of original data available at
/gpfs/fs001/cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/curation/code/sandbox/validator_outputs/d2_r2_validation.csv. Data backed up to datalad.
BIDS Optimization:
*NOTE: any files removed from curation/BIDS dataset noted in /cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/curation/curation_*_cmd.sh scripts, which are written by cubids-purge. Any files renamed (Acquisition Variants) noted in /cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/curation/code/iterations/apply*_cmd.sh scripts, which are written by cubids-apply.
- BIDs groups from Iteration 1.2 reviewed by Ted and Tinashe
- reviewed subject files for duplicates, no subj with more than one T1w or each type of fmap (phase1, phase2, magnitude1, magnitude2)
- 118 subj have full set of phase1&2, magnitude1&2 files
- 5 subj have only phasediff files (mislabeled phase1) but no magnitude files
- 1 subj has only phase1 & phase2 files but no magnitude files
- identified 3 subjects who have T2 data (KeyParamGroup=datatype-anat_suffix-T2w__1) in addition to T1 that compromise AcqGroup 3
- Plan to add A/B designation task entity for files to disambiguate task version (cardA,cardB, faceA, or faceB) performed during each run. Data currently contained in SeriesDescription, see task-match.ipynb *counts using validator_err_counts.ipynb *
- Iteration 2
- made sure all files in curation/BIDS checked into datalad
- T2 files to be removed written to code/sandbox/T2w.txt using validator_err_counts.ipynb, ran
cubids-purge:cubids-purge --use-datalad /cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/curation/BIDS /cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/curation/code/sandbox/T2w.txt - fmap files to be removed written to
Margaret/Day2/curation/code/sandbox/validator_outputs/iteration1.2/fmap_to_rm.txtusing validator_err_counts.ipynb, rancubids-purge:cubids-purge --use-datalad /cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/curation/BIDS /cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/curation/code/sandbox/validator_outputs/iteration1.2/fmap_to_rm.txt - Reran
cubids-validatoriter2 with--ignore_nifti_headersand--ignore_subject_consistencyflags; outputs identical to Iteration 1.2 above (reviewed using validator_parser.ipynb). - Reran
cubids-group- still resulted in 23 acquisition groups, including addition of 4 new KeyParamGroups (reviewed using group_compare.ipynb): acquisition-VARIANTNoFmap_datatype-func_run-2_suffix-bold_task-card acquisition-VARIANTNoFmap_datatype-func_run-2_suffix-bold_task-face acquisition-VARIANTNoFmap_datatype-func_run-1_suffix-bold_task-face acquisition-VARIANTObliquityNoFmap_datatype-func_suffix-bold_task-rest acquisition-VARIANTNoFmap_datatype-func_suffix-bold_task-rest - Groupings approved by Ted and Tinashe, ran
cubids-applywithout modifications to iter2_summary or iter2_files:cubids-apply --use-datalad BIDS code/iterations/iteration2/iter2_summary.csv code/iterations/iteration2/iter2_files.csv code/iterations/apply1 cubids-applycreated apply1_full_cmd.sh (renamed to apply1a_full_cmd.sh) but unsuccessful in renaming files; internet disconnected and wasn’t able to copy error from jupyter terminal, reran command and reproduced error:raise CommandError( datalad.support.exceptions.CommandError: CommandError: 'bash code/iterations/apply1_full_cmd.sh' failed with exitcode 127 under /gpfs/fs001/cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/curation/BIDS'- decided to edit iter2_summary.csv and rerun per Tinashe’s request to rename lengthy T1w keygroups, then will try to solve cubids-apply error
- renamed:
- KeyParamGroup datatype-anat_suffix-T1w__3 to acquisition-VARIANTAllwithParallelReductionFactorInPlane_datatype-anat_suffix-T1w
- KeyParamGroup datatype-anat_suffix-T1w__4 to acquisition-VARIANTAll_datatype-anat_suffix-T1w
Rancubids-applywith above modifications to iter2_summary.csv:cubids-apply --use-datalad BIDS code/iterations/iteration2/iter2_summary.csv code/iterations/iteration2/iter2_files.csv code/iterations/apply2
- renamed:
- Iteration 3
- per Sydney Covitz’s recommendations, reran
cubids-group usingfull paths:cubids-group --use-datalad /cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/curation/BIDS /cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/curation/code/iterations/iteration3/iter3 - resulted in 23 acquisition groups, including addition of 4 new KeyParamGroups (reviewed using group_compare.ipynb): acquisition-VARIANTNumVolumesNoFmap_datatype-func_run-2_suffix-bold_task-face acquisition-VARIANTNumVolumesNoFmap_datatype-func_run-1_suffix-bold_task-face acquisition-VARIANTNumVolumesNoFmap_datatype-func_suffix-bold_task-rest acquisition-VARIANTNumVolumesNoFmap_datatype-func_suffix-bold_task-rest
- reviewed with Sydney, discovered that prior
cubids-applyattempts had succcessfully renamed IntendedFors field in fmap json’s but exited before being able to rename the filenames (due to the fact that the files.csv had the /gpfs/fs001/ string in it because cubids-group was run using relative paths), resulting in “NoFmap” additions above. Per Sydney’s recommendation runningcubids-undoto un-rename IntendedFors; rerancubids-groupand finallycubids-applyusing abs. paths.- ran
git clean -f -dto remove untracked changes in .ipynb_checkpoints - ran
cubids-undo, used intendedfor_rename.ipynb to verify once VARIANT renames had been cleared. Datalad executes undone tracked below:- HEAD is now at 69e473c Renamed IntendedFors
- HEAD is now at 1ccd650 Renamed IntendedFors
- HEAD is now at c8466c7 Renamed IntendedFors
- HEAD is now at abc67c1 Renamed IntendedFors
- HEAD is now at 26b23ee Renamed IntendedFors
- HEAD is now at edfb983 updating .ipynb
- ran
- per Sydney Covitz’s recommendations, reran
- Iteration 4
- successfully removed all VARIANT intendedfors, rerunning:
cubids-group --use-datalad /cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/curation/BIDS /cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/curation/code/iterations/iteration4/iter4 - reviewed groupings against iter2 using group_compare.ipynb, no changes. Renamed the lengthy T1w keygroups per Tinashe’s request:
- datatype-anat_suffix-T1w__3 : acquisition-VARIANTAllwithParallelReductionFactorInPlane_datatype-anat_suffix-T1w
- datatype-anat_suffix-T1w__4 : acquisition-VARIANTAll_datatype-anat_suffix-T1w
- ran:
cubids-apply --use-datalad /cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/curation/BIDS /cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/curation/code/iterations/iteration4/iter4_summary.csv /cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/curation/code/iterations/iteration4/iter4_files.csv /cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/curation/code/iterations/apply2 cubids-applysuccessful- ran
cubids-validate, no new errors or warnings: EVENTS_TSV_MISSING : 495 scans README_FILE_MISSING : 1 count NO_AUTHORS : 1 count
- successfully removed all VARIANT intendedfors, rerunning:
- Iteration 5
- 3 exemplar subjects (sub-15546, sub-16181, & sub-12235) failed running fmriprep due to abberant image shape (64, 64, 43) in fmap images. Each subject compromised a unique Acquisition group. Deleting all fmap images (listed using Dim3_err_fmaps.ipynb):
cubids-purge --use-datalad /cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/curation/BIDS /cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/curation/code/sandbox/fmap_to_rm2.txt - ran
cubids-group, new groups ID’d for above subj (NoFMap) that will be merged into existing NoFMap groups withcubids-apply - ran
cubids-applywithout changes with prefix apply3, successful - ran
cubids-validate, parsed using validator_parser.ipynb, no new errors or warnings: EVENTS_TSV_MISSING : 495 scans README_FILE_MISSING : 1 count NO_AUTHORS : 1 count
- 3 exemplar subjects (sub-15546, sub-16181, & sub-12235) failed running fmriprep due to abberant image shape (64, 64, 43) in fmap images. Each subject compromised a unique Acquisition group. Deleting all fmap images (listed using Dim3_err_fmaps.ipynb):
- Iteration 6
- 3 subjects (sub-13373, sub-14858, sub-15709) failed fmriprep due to CRC error, deleting nifti files identified in log outputs:
cubids-purge --use-datalad /cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/curation/BIDS /cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/curation/code/sandbox/CRC_err_to_rm.txt*NOTE: these 3 scans can be rerun in the future with a different version of fmriprep that doesn’t have this gzip error! - ran
cubids-group, no new variants (no RenameKeyGroups for non-fmap KeyGroups) - Tinashe reviewed, no need forcubids-apply/validate
- 3 subjects (sub-13373, sub-14858, sub-15709) failed fmriprep due to CRC error, deleting nifti files identified in log outputs:
- Timing Files
- created event timing files (events.tsv) based on
K23_fmri_paradigm.xlsprovided by Dan Wolf- used stick files to create two .csv’s listing all events for run-1 (task A) and run-2 (task B) respectively (cardA and faceA have the same timings/outcome order, just the stimuli are different; cardB and faceB have the same timings/outcomes)
- ONSET TIMES IN STICK FILES REFLECT FACT THAT ANALYSIS PIPELINE DELETED FIRST 20 SECONDS=10TR OF BOLD RUNS, DURING WHICH TWO “DUMMY” TASK TRIALS OCCURRED
- converted to .tsv’s using csv_to_tsv.ipynb
- copied .tsvs into Day2/curation/BIDS, reran
cubids-validate; took several iterations of renaming events.tsv’s so they will be correctly applied/pass validator, succeded on iteration 5 (/cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/curation/code/sandbox/validator_outputs/tsv5_validation.csv)
- created event timing files (events.tsv) based on
Preprocessing Pipelines
- fMRIPrep (version 20.2.3)
- Margaret Gardner is responsible for running preprocessing pipelines/audits on CUBIC
- Exemplar Testing:
- ran
cubids-copy-exemplars --use-datalad /cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/curation/BIDS /cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/testing/exemplars_dir /cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/curation/code/iterations/apply2_AcqGrouping.csv - Path to exemplar dataset (annexed to datalad):
/cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/testing/exemplars_dir - Path to fmriprep container (.sif copied from dropbox, annexed to datalad):
/cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/testing/exemplars_test/fmriprep-container - ran
(tail -n 1 code/qsub_calls.sh)w/out modifications to participant_job.sh or fmriprep_zip.sh but no output branch created and didn’t save job number; reran, job writing to analysis/logs but seems unable to create new datalad branch (pushingitremote... line 32: datalad: command not found); ** editedparticipant_job.shto correct conda environment (from base to margaret_reward) and run job in /cbica/comp_space; failed b/c had comments in-line onfmriprep_zip.sh** reviewed with Tinashe and editedfmriprep_zip.sh; reran job 1424461 (“fpsub-12583”) has been submitted - completed successfully ** ranbash code/qsub_calls.sh, submitted jobs 1679260 through 1679282 & merged to merge_ds (with help from Sydney & Matt - issues with merge failing since test sub-12583 had already been merged, followed their instruction to delete both sub-12583 branches since .zip files already present in merge_ds) - error in sub-15546, sub-16181, & sub-12235 (fmap images with Dim3Size=43, unable to construct fmaps - removing all fmap images for these subjects, see Iteration 5 above) *Note: flag
--use-syn-sdcnot included in fmriprep run call, so no susceptibility distortion correction was run for subjects without fieldmaps - sub-12583 (test sub) doesn’t have branch in output_ria but is fine in audit
- Path to exemplar outputs: /cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/testing/fmriprep/output_ria testing dir deleted to save space on CUBIC on 12/2/21, once production completed
- ran
- Production:
- ran
qsub_calls.sh, submitted jobs 1831777 through 1831903 - only 84 files in logfile, 123 branches created under output_ria
- running
merge_outputs.shand fmriprep-audit to identify failed subj - edited
concat_outputs.sh(still old version on github) to pull tinashe’s newconcatenator.pyedits and edited line 12 ‘concat_ds/csvs’ to ‘csvs’
- running
- reviewed
FMRIPREP_AUDIT.csv, 4 subj failed:- sub-13373 - nipype.workflow ERROR: Node bold_to_std_transform.a0 failed to run on host 2119fmn002… File “indexed_gzip/indexed_gzip.pyx”, line 635, in indexed_gzip.indexed_gzip._IndexedGzipFile.seek indexed_gzip.indexed_gzip.CrcError: CRC/size validation failed - the GZIP data might be corrupt
- used
gzip -t -vto validate CRC size for sub-13373_ses-day2_task-face_run-01_bold.nii.gz, OK
- used
- sub-14858 - same as above, err on sub-14858_ses-day2_task-card_acq-VARIANTNoFmap_run-02_bold.nii.gz
- sub-15709 - same as above, err on sub-15709_ses-day2_task-rest_bold.nii.gz
- sub-17113 - no error message, log o and e incomplete - to rerun
- removing scans with CRC error, see Iteration 6 above - pushed BIDS updates to input_ria, rerunning qsub calls for sub-13373 (job 1974456), sub-14858 (job 1974459), sub-15709 (job 1974460), and sub-17113 (job 1974462); ran successfully based on logs, deleted merge_ds and reran
merge_outputs.sh. Regot and reran fmriprep-audit
- sub-13373 - nipype.workflow ERROR: Node bold_to_std_transform.a0 failed to run on host 2119fmn002… File “indexed_gzip/indexed_gzip.pyx”, line 635, in indexed_gzip.indexed_gzip._IndexedGzipFile.seek indexed_gzip.indexed_gzip.CrcError: CRC/size validation failed - the GZIP data might be corrupt
- all 125 subjecs successfully processed
- Path to production inputs:
/cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/curation/BIDS - Path to fmriprep run command:
/cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/production/fmriprep/analysis/code/fmriprep_zip.sh - Path to production outputs:
/cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/production/fmriprep/output_ria - Path to fmriprep production audit:
/cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/production/fmriprep-audit/FMRIPREP_AUDIT.csv - Path to freesurfer production audit:
/cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/production/freesurfer-audit** plotted Euler numbers generated by freesurfer_audit and plotted distribution. Sub-15433 recommended to be excluded from subsequent analyses (Euler=782). Reviewed sub-11305 (238) and sub-11399 (224) with Ted but ok’d
- ran
- xcp-abcd
- Production:
- edited participant_job.sh, xcp_zip.sh to update python environment, update “xcp-abcd-0-0-4” to “xcp-abcd-0-0-8” (matching container name and Tinashe’s scripts for other Reward projects)
- ran test subject job 203853 (“xcpsub-17838”), successful!
- submitted remaining jobs, successful!
- submitted qsub_calls.sh for xcp-audit
- wget and running bootstrap-quickunzip.sh to clone/unzip xcp outputs to xcp-derivatives; something didn’t work, seemed to overwrite unzip.sh?
- removed and wgot again, but had typo in path to xcp dir, rerunning with corrected path:
qsub -cwd -N "d2_unzip" bootstrap-quickunzip.sh /cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/production/xcp- job 213392 (“d2_unzip”) has been submitted; job didn’t seem to run, no outputs; see e and o output files. Rerunning from terminal (not qsub-ed), renamed dirwolf_satterthwaite_rewardtoderivatives-unzipped - concatenated
*space-MNI152NLin6Asym_desc-qc_res-2_bold.csvoutputs with xcp_qc_concat.ipynb, plotted and saved outputs to github dir qc_plots
- removed and wgot again, but had typo in path to xcp dir, rerunning with corrected path:
- Path to production inputs:
/cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/production/fmriprep/merge_ds - Path to xcp run command:
/cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/production/xcp/analysis/code/xcp_zip.sh - Path to production outputs:
/cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/production/xcp/output_ria - Path to xcp production audit:
/cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/production/xcp-audit/XCP_AUDIT.csv - Path to xcp derivatives:
/cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/production/derivatives-unzipped/DERIVATIVES/XCP - Path to xcp derivatives (concatenated):
/cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/curation/code/sandbox/qc_d2.csv
- Production:
Post Processing
- Who is using the data/for which projects are people in the lab using this data?
- Link to project page(s) here
- For each post-processing analysis that has been run on this data, fill out the following
- Who performed the analysis?
- Where it was performed (CUBIC, PMACS, somewhere else)?
- GitHub Link(s) to result(s)
- Did you use pennlinckit?
- https://github.com/PennLINC/PennLINC-Kit/tree/main/pennlinckit
- FEAT task analysis
- fun side-quest for personal growth run by Margaret Gardner on CUBIC
- wrote .txt timing files using fsl_timing_create.sh
- Dan provided original feat analysis files for reference, saved under
fsl_sandbox/dan_orig- “the events folder has all the stickfiles, lots of different variations. the feat directory has a feat directory for this control participant’s cardA analysis: 11242_03360; the nifti images is that persons 4D bold timeseries used for that feat analysis.”
- running on raw data from 3 subj randomly selected from Acquisition Group 1 (sub-16291, sub-15732, & sub-15761) in
/cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/fsl_sandboxto familiarize with fsl workflow before adapting to accomodate fmriprep outputs; scripts run from git repo directoryfsl- ran BET on sub-15732 with default settings, pial surface not fully removed - reran with f=0.7 but removed too much, sticking with default f=0.5
- running FEAT preprocessing on sub-15732 card run-01: deleting 10 vol, set smoothing to 6.0
- error in Registration: Could not find a supported file with prefix “/gpfs/fs001/cbica/projects/wolf_satterthwaite_reward/Margaret/Day2/fsl_sandbox/BIDS/card_run-01.feat/example_func.nii.gz”
- talked to Greer and discovered error was in bet outputting .hdr/.imgs instead of .nii.gz - need to define FSLOUTPUTTYPE=NIFTI_GZ. Removed all fsl outputs/reverting to raw BIDs to run again
- ran BET on sub-15732 with default settings (-f 0.5), extraction looks good
- ran FEAT preprocessing on sub-15732 card run-01: deleting 10 vol, set smoothing to 6.0; successful,
- ran Stats on sub-15732 card run-1 with 4 EVs (cue, anticipation, win, lose) and 3 contrasts: (0, 0, 1, 0); (0, 0, 0, 1); (0, 0, 1, -1)
- running full analyses on sub-15732 card run-1 with 4 EVs (cue, anticipation, win, lose) and 3 contrasts: (0, 0, 1, 0); (0, 0, 0, 1); (0, 0, 1, -1); successful, removed old feat directories (preprocessing/stats only)
- duplicating/editing design.fsf to github, create seperate design.fsf’s for each task/run combo (starting with card1, potentially iterate across card/task in the future since they have identical EVs)
- testing
design_card1.fsfon sub-16291, ran successfully - created design files for each task/run and updated run_1stLevel_Analysis.sh
- ran run_1stLevel_Analysis.sh note: outputs and QCs not reviewed since this was for an exercise - would review and/or rerun if intending to use outputs in the future
- removed sub-16291/card1+.feat dir
- running 2nd level fixed-effects for card task (averaged across run 1 and 2 for each subj), output to
fsl_sandbox/card_2ndLevel.gfeat - running 3rd level FLAME 1 model for card cope 3 (win-lose), default thresholds (cluster z=3.1, p=0.05); inputs ``fsl_sandbox/card_2ndLevel.gfeat/cope3.feat/stats/cope?.nii.gz
; output tofsl_sandbox/card_3rdLevel_win-lose.gfeat`- also ran uncorrected analysis to
fsl_sandbox/card_3rdLevel_win-lose_uncorr.gfeatjust for fun
- also ran uncorrected analysis to
To Do
- backup to PMACS
- rename task entity (1 and 2 vs A and B)
- script group level task analyses in FEAT
- bootstrap FEAT analyses for datalad
- adapt feat script to accept fmriprep outputs