HIERARCHICAL NEURODEVELOPMENT AND THE SENSORIMOTOR-ASSOCIATION AXIS
The human brain undergoes a prolonged period of cortical development that spans multiple decades. During childhood and adolescence, cortical development progresses from lower-order, primary and unimodal cortices with sensory and motor functions to higher-order, transmodal association cortices subserving executive, socioemotional, and mentalizing functions. The spatiotemporal patterning of cortical maturation thus proceeds in a hierarchical manner, conforming to an evolutionarily rooted, sensorimotor-to-association axis of cortical organization. This developmental program has been characterized by data derived from multimodal human neuroimaging and is linked to the hierarchical unfolding of plasticity-related neurobiological events. Critically, this developmental program serves to enhance feature variation between lower-order and higher-order regions, thus endowing the brain’s association cortices with unique functional properties. However, accumulating evidence suggests that protracted plasticity within late-maturing association cortices, which represents a defining feature of the human developmental program, also confers risk for diverse developmental psychopathologies.
Review article published in Neuron (2021) as Neurodevelopment of the association cortices: Patterns, mechanisms, and implications for psychopathology
Publication DOI
https://doi.org/10.1016/j.neuron.2021.06.016
Project Lead
Valerie J. Sydnor
Faculty Lead
Theodore D. Satterthwaite
Collaborators
Bart Larsen, Dani S. Bassett, Aaron Alexander-Bloch, Damien A. Fair, Conor Liston, Allyson P. Mackey, Michael P. Milham, Adam Pines, David R. Roalf, Jakob Seidlitz, Ting Xu, Armin Raznahan
Github Repository
https://github.com/PennLINC/S-A_ArchetypalAxis
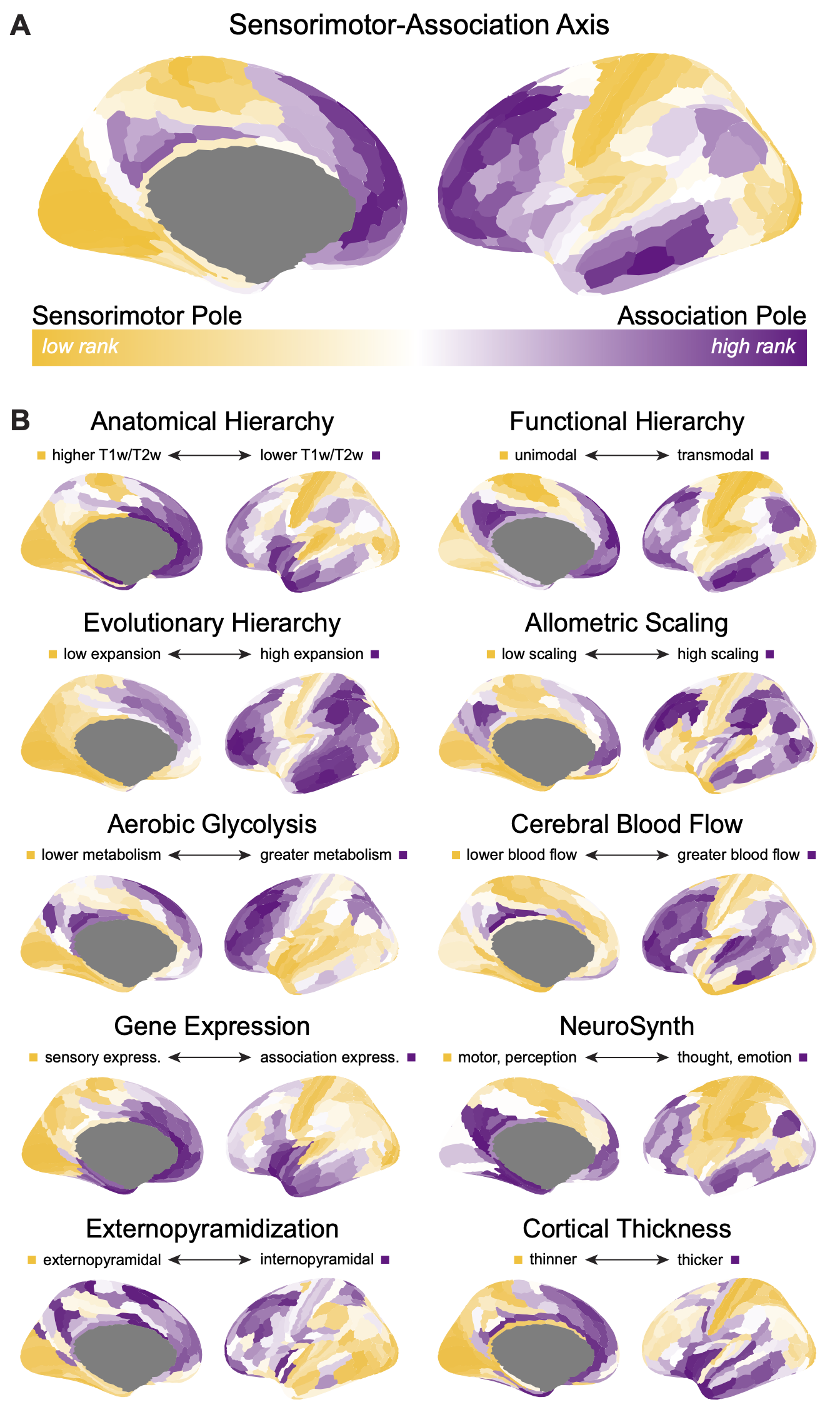
THE SENSORIMOTOR-ASSOCIATION AXIS
The 10 cortical feature maps and the archetypal sensorimotor-association (S-A) axis displayed in Figures 2B and 2A (respectively) of Sydnor et al., 2021, Neuron are available here in Glasser360-MMP and Schaefer400-17Network parcellations, as well as in FSLR (HCP) and fsaverage5 vertices.
Raw data for the 10 cortical features can be found in the associated github repo in $atlas/brainmaps_$atlas.csv. These are macrostructural, microstructural, functional, metabolic, transcriptomic, and evolutionary features that exhibit systematic variation between lower-order primary sensorimotor regions and higher-order transmodal association regions. If you use data from these feature maps, please be sure to cite the original data source(s)+.
The rank ordering of each cortical area along the archetypal S-A axis can be found in the repo in $atlas/Sensorimotor_Association_Axis_AverageRanks.csv; please cite the review if you use the S-A axis in your work.
Below is a description of all files in PennLINC’s S-A_ArchetypalAxis repo:
GLASSER360_MMP
Includes code used to generate Figure 2A-D in Sydnor et al., 2021, Neuron.
- brainmaps.Rmd: code to visualize brain maps for the 10 cortical features (Figure 2B), the archetypal S-A axis (Figure 2A), and S-A axis rank variability (Box 1 Figure), and to compute and visualize sensorimotor and association tertile z-scores (Figure 2D)
- spintests.Rmd: code to generate a Spearman’s rank correlation matrix for the 10 cortical features, and to quantify correlation significance using a parcel-based spatial permutation spin test (pspin) (Figure 2C)
- brainmaps_glasser.csv: the 10 cortical features displayed in Figure 2B in 180 left hemisphere Glasser parcels
- glasser_regions.csv: list of left hemisphere Glasser parcel names (rows correspond to rows in brainmaps_glasser.csv)
- Sensorimotor_Association_Axis_AverageRanks.csv: each parcel’s average ranking (raw mean) and final ranking (re-ranked average rank) along the S-A axis. Average rankings were obtained by averaging individual parcel rankings across the 10 cortical features. Average rankings were re-ranked from 1-180 to derive the archetypal S-A axis (displayed in Figure 2A)
SCHAEFER400_17NETWORK
Includes code to generate cortical maps in the Schaefer400 atlas.
- brainmaps.Rmd: code to extract measures for the 10 cortical features and to derive the archetypal S-A axis
- brainmaps_schaefer.csv: the 10 cortical features displayed in Figure 2B in Schaefer400 parcels. (Note, here the histological measure Externopyramidization is replaced by the updated BigBrainWarp histological gradient Hist-G2)
- schaefer400_regions.csv: list of Schaefer400 17 network parcel names (rows correspond to rows in brainmaps_schaefer.csv)
- Sensorimotor_Association_Axis_AverageRanks.csv: the S-A axis in Schaefer400. Average rankings and final rankings are provided for the whole brain (ranked 1-400) and for left and right hemispheres individually (ranked 1-200)
FSLRVertex
Includes code to generate cortical maps in fslr 32k vertices (HCP surface space). In the fslr surface, each cortical hemisphere has a total of 32492 vertices, including the medial wall. Excluding medial wall vertices, the left cortex has 29696 vertices and the right cortex has 29716 vertices. Medial wall vertices are identified in medialwall.mask.$hemicortex.csv and correspond to the default HCP fslr 32k mesh medial wall. S-A axis rankings for each vertex are provided in the cifti format (SensorimotorAssociation_Axis.dscalar.nii) and in gifti format (SensorimotorAssociation_Axis_$H.fslr32k.func.gii) as well as in a csv.
- brainmaps.Rmd: code to extract measures for the 10 cortical features and to derive the archetypal S-A axis at the vertex level
- brainmaps_fslr.csv: the 10 cortical features displayed in Figure 2B in fslr surface vertices. The data in each column was extracted from the identically named $feature.dscalar.nii file provided in this repo. (Note, here the histological measure Externopyramidization is replaced by the updated BigBrainWarp histological gradient Hist-G2). This csv contains a total of 59412 rows; rows 1:29696 correspond to the left cortex, rows 29697:59412 correspond to the right cortex
- Sensorimotor_Association_Axis_AverageRanks.csv: the S-A axis in fslr surface vertices. Average rankings (“averagerank” columns) and final rankings (“finalrank” columns) are provided for the whole brain (ranked 1-59412) and for left and right hemispheres individually. Z-scored whole brain rankings are also provided in the finalrank.wholebrain.zscore column. This csv contains a total of 59412 rows; rows 1:29696 correspond to the left cortex, rows 29697:59412 correspond to the right cortex
- SensorimotorAssociation_Axis.dscalar.nii: S-A axis rankings (finalrank.wholebrain) provided in cifti file format
- SensorimotorAssociation_Axis_LH.fslr32k.func.gii: left hemisphere S-A axis rankings (finalrank.wholebrain) provided in gifti metric file format
- SensorimotorAssociation_Axis_RH.fslr32k.func.gii: right hemisphere S-A axis rankings (finalrank.wholebrain) provided in gifti metric file format
- SensorimotorAssociation_Axis_parcellated: this folder contains parcellated versions of the vertex-wise fslr S-A axis (pscalar.nii files) and the corresponding atlas_dlabel_files used for parcellation
FSaverage5
Includes code to resample the fslr 32k S-A axis to the fsaverage5 10k surface. In the fsaverage5 surface, each cortical hemisphere has a total of 10242 vertices, including the medial wall.
- fslr_to_fsaverage5.sh: code to resample S-A axis whole brain rankings from fslr vertices to fsaverage5 vertices using wb_command -cifti-separate and -metric-resample
- SensorimotorAssociation_Axis_LH.fsaverage5.func.gii: left hemisphere S-A axis rankings in fsaverage5, provided in gifti metric file format
- SensorimotorAssociation_Axis_RH.fsaverage5.func.gii: right hemisphere S-A axis rankings in fsaverage5, provided in gifti metric file format
+
Anatomical Hierarchy: Glasser and Van Essen, 2011, J Neurosci
Functional Hierarchy: Margulies et al., 2016, PNAS
Evolutionary Hierarchy: Hill et al., 2010, PNAS
Allometric Scaling: Reardon, Seidlitz, et al., 2018, Science
Aerobic Glycolysis: Vaishnavi et al., 2010, PNAS; Glasser et al., 2015, NeuroImage
Cerebral Blood Flow: Satterthwaite et al., 2014, PNAS
Gene Expression: Burt et al., 2018, Nat Neurosci
NeuroSynth: Yarkoni et al., 2011, Nat Methods
Externopyramidization: Paquola et al., 2020, PLoS Biol / HIST-G2: Paquola et al., 2021, eLife
Cortical Thickness: Glasser et al., 2016, Nature