Project Information and Code Reproducibility Guide
Sex Differences in Functional Topography of Association Networks
Prior work has shown that there is substantial interindividual variation in the spatial distribution of functional networks across the cerebral cortex, or “functional topography”. However, it remains unknown whether there are normative sex differences in the topography of individualized networks. Here we leverage regularized non-negative matrix factorization to define individualized functional networks in 693 youth ages 8-23y imaged with fMRI as part of the Philadelphia Neurodevelopmental Cohort. Support vector machines were applied to examine sex differences in multivariate patterns of functional topography. Generalized additive models with penalized splines were then used to examine the impact of sex on topography at a more granular level. We used chromosomal enrichment analyses to assess the correlation between gene expression (Allen Human Brain Atlas) and sex differences in functional topography, evaluating significance with gene-wise non-parametric permutation tests. This project identifies normative sex differences in the functional topography of association networks and highlight the role of sex as a biological variable in shaping functional topography.
Project Lead
Sheila Shanmugan
Faculty Lead
Theodore D. Satterthwaite
Analytic Replicator
Zaixu Cui (imaging)
Jakob Seidlitz (genetics)
Max Bertolero (Figure 2b)
Collaborators
Jakob Seidlitz, Zaixu Cui, Azeez Adebimpe, Danielle S. Bassett, Maxwell A. Bertolero, Christos Davatzikos, Damien A. Fair, Raquel E. Gur, Ruben C. Gur, Hongming Li, Adam Pines, Armin Raznahan, David R. Roalf, Russell T. Shinohara, Jacob Vogel, Daniel H. Wolf, Yong Fan, Aaron Alexander-Bloch
Project Start Date
January 2019
Current Project Status
In preparation
Datasets
PNC
Github Repository
https://github.com/sheilashanmugan/funcParcelSexDiff1
Path to Data on Filesystem
/cbica/projects/funcParcelSexDiff/data
Publication DOI
Conference Presentations
CODE DOCUMENTATION
The steps below detail how to replicate this project, including statistical analysis and figure generation.
Part 1: Atlas Generation and Network Parcellation
-
Sample selection, atlas generation, and individual network parcellation.
These steps were completed as part of prior work (Cui et al., 2020) using scripts located at https://github.com/ZaixuCui/pncSingleFuncParcel
Loading matrices for each of the 693 subjects used in this project can be found here: /cbica/projects/funcParcelSexDiff/data/Revision/SingleParcellation/SingleAtlas_Analysis/FinalAtlasLoading
The following steps use this preprocessed data.
-
Create violin plot in Figure 2b with atlasLoadingScripts/diceOverlapn693_17net_20220506.R
This script creates a violin plot of the Dice coefficient between the group atlas and each subject for all 17 networks.
Part 2: Multivariate Pattern Analysis
-
Add /matlabFunctions/Toolbox/Code_mvNMF_l21_ard_v3_release/ to matlab path.
This directory contains functions called in subsequent steps
-
Add /SVM_scripts/scripts_from_zaixu to matlab path.
This directory contains functions called in subsequent steps
-
Run /SVM_scripts/makeNonZeroMatrix.m to make nonzero index.
This script creates the non-zero index needed to visualize results
-
Run SVM with /SVM_scripts/run_SVM/SVM_sex_2fold_CSelect_Cov_SubIndex_Perm_20200719.m
This frist part of this script (100 Repeat) submits 100 jobs, each of which are one of the 100 repetitions of SVM predictions. The second part of this script (Permutation, 1000 times) submits 1000 jobs, each of which are one of 1000 permutations that will be used for significane testing of accuracy. In each run, sex is permuted across the training subset without replacement.
-
Create files for workbench visualization by running /SVM_scripts/Weight_Visualize_Workbench/Weight_Visualize_Workbench_Sex_SVM2foldCSelectCovSubIndex_20200721.m
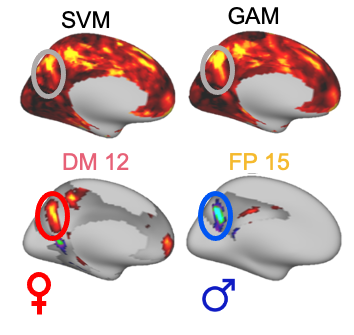
This script creates files for workbench visualization. It creates ciftis that display the weight of the first 25% regions with the highest absolute weight from SVM (Figure 3C). It also creates a cift that displays the sum of the absolute value of weight across the 17 networks (Figure 3D).
-
Visualize files in workbench (Figures 3C and 3D)
/Applications/workbench/bin_macosx64/wb_view /Users/sheilash/Desktop/projects/pfn_sex_diff/inputData/spec_files/rh.inflated.surf.gii /Users/sheilash/Desktop/projects/pfn_sex_diff/inputData/spec_files/lh.inflated.surf.gii /cbica/projects/funcParcelSexDiff/results/PredictionAnalysis/SVM/2fold_CSelect_Cov_SubIndex/Sex_CovAgeMotion/Permutation/res_MultiTimes/AtlasLoading/WeightVisualize_Sex_SVM_2fold_CSelect_Cov_MultiTimes/First25Percent/w_Brain_Sex_First25Percent_Network_9.dscalar.nii /Users/sheilash/Desktop/projects/pfn_sex_diff/inputData/Group_Loading_17Networks/Group_AtlasLoading_Network_9.dscalar.nii &
> /Applications/workbench/bin_macosx64/wb_view /Users/sheilash/Desktop/projects/pfn_sex_diff/inputData/spec_files/rh.inflated.surf.gii /Users/sheilash/Desktop/projects/pfn_sex_diff/inputData/spec_files/lh.inflated.surf.gii /cbica/projects/funcParcelSexDiff/results/PredictionAnalysis/SVM/2fold_CSelect_Cov_SubIndex/Sex_CovAgeMotion/Permutation/res_MultiTimes/AtlasLoading/WeightVisualize_Sex_SVM_2fold_CSelect_Cov_MultiTimes/First25Percent/w_Brain_Sex_First25Percent_Network_12.dscalar.nii /Users/sheilash/Desktop/projects/pfn_sex_diff/inputData/Group_Loading_17Networks/Group_AtlasLoading_Network_12.dscalar.nii &
> /Applications/workbench/bin_macosx64/wb_view /Users/sheilash/Desktop/projects/pfn_sex_diff/inputData/spec_files/rh.inflated.surf.gii /Users/sheilash/Desktop/projects/pfn_sex_diff/inputData/spec_files/lh.inflated.surf.gii /cbica/projects/funcParcelSexDiff/results/PredictionAnalysis/SVM/2fold_CSelect_Cov_SubIndex/Sex_CovAgeMotion/Permutation/res_MultiTimes/AtlasLoading/WeightVisualize_Sex_SVM_2fold_CSelect_Cov_MultiTimes/w_Brain_Sex_Abs_sum.dscalar.nii &
-
Calculate summary statistics with SVM_scripts/calc_accuracy/average_acc_sens_spec_SVM_multiTimes_20200720.m
This script aggregates accuracy, sensitivity, and specificity across the 100 SVM repeats then averages them. It then compares this accuracy to permuted accuracies to calculate a p-value for accuracy. Save
acc_AllModels1_permasSVM_perm_accuracy.csv. This is used as the input when generating the histogram inset in step 10 below. -
Get y values needed to draw ROC with SVM_scripts/roc/yvalues_100_20201108.R
This script aggregates the Y values across the 100 SVM repeats. The output of this step is used in step 9 below.
-
Create ROC with SVM_scripts/roc/SVM_ROC.m
This script uses the function /SVM_scripts/roc/AUC_Calculate_ROC_Draw2.m to create the plot in Figure 3a.
-
Create histogram inset in figure 3a with SVM_scripts/roc/SVM_accuracy_histogram.R
This script creates a histogram of accuracies from the permutation test.
-
Create barplot of SVM Weights with SVM_scripts/barplots/sums_weights_stackedBarplot_20201021.R
This script creates the plot in Figure 3b
Part 3: Univariate approach
-
Submit atlasLoadingScripts/sexEffect_atlasLoading_20200612.R to qsub to calculate effect of sex on atlas loadings
This script aggregates atlas loadings for all subjects, runs a GAM at each vertex to determine the effect of sex while controlling for age and motion, then corrects for multiple comparisons.
qsub -l h_vmem=10.5G,s_vmem=10.0G /cbica/projects/funcParcelSexDiff/scripts/run_R_script.sh /cbica/projects/funcParcelSexDiff/scripts/atlasLoadingScripts/sexEffect_atlasLoading_20200612.R -
Write effect map for each network with WriteEffectMap/WriteEffectMap_Workbench_Sex_20200712.m
This script takes the output of the GAMs from step 1 and creates the CIFTI files of the effect of sex at each vertex.
-
Create Figure 4A with WriteEffectMap/WriteEffectMap_Workbench_Sex_AbsSum_20200712.m
This script sums the absolute value of the effect of sex across all 17 networks and creates a CIFTI file of this effect.
-
Visualize files in workbench (Figure 4A, 4D, and 4E)
/Applications/workbench/bin_macosx64/wb_view /Users/sheilash/Desktop/projects/pfn_sex_diff/inputData/spec_files/rh.inflated.surf.gii /Users/sheilash/Desktop/projects/pfn_sex_diff/inputData/spec_files/lh.inflated.surf.gii /cbica/projects/funcParcelSexDiff/results/GamAnalysis/AtlasLoading/SexEffects/UnthreshAbsSum/Gam_Sex_Abs_sum.dscalar.nii &
/Applications/workbench/bin_macosx64/wb_view /Users/sheilash/Desktop/projects/pfn_sex_diff/inputData/spec_files/rh.inflated.surf.gii /Users/sheilash/Desktop/projects/pfn_sex_diff/inputData/spec_files/lh.inflated.surf.gii /Users/sheilash/Desktop/projects/pfn_sex_diff/inputData/Group_Loading_17Networks/Group_AtlasLoading_Network_9.dscalar.nii /cbica/projects/funcParcelSexDiff/results/GamAnalysis/AtlasLoading/SexEffects/Gam_Z_FDR_Sig_Vector_All_Sex_Network_9.dscalar.nii &
/Applications/workbench/bin_macosx64/wb_view /Users/sheilash/Desktop/projects/pfn_sex_diff/inputData/spec_files/rh.inflated.surf.gii /Users/sheilash/Desktop/projects/pfn_sex_diff/inputData/spec_files/lh.inflated.surf.gii /Users/sheilash/Desktop/projects/pfn_sex_diff/inputData/Group_Loading_17Networks/Group_AtlasLoading_Network_12.dscalar.nii /cbica/projects/funcParcelSexDiff/results/GamAnalysis/AtlasLoading/SexEffects/Gam_Z_FDR_Sig_Vector_All_Sex_Network_12.dscalar.nii &
-
Aggregate group level matricies with WriteEffectMap/barplots/Make_SexEffects_mat_gam_FDR_sig.m
This script aggregates group level matricies for each network that denote significant verticies. The output of this step is the input for step 6.
-
Create barplot in Figure 4C with WriteEffectMap/barplots/sum_gam_barplot_20210417.R
This script creates a barplot of the number of verticies that survives FDR correction for each network.
Part 4: Spin test to compare results from multivariate pattern analysis (Figure 3D) and GAMs (Figure 4A)
-
Download and add spintest/scripts/ to matlab path.
This directory contains functions called in subsequent steps. These functions were originally downloaded from here
-
Prepare data for spin test with spintest/prepare_data_for_spintest_20201104.R
This script formats data for the spin test.
-
Run spin test with spintest/spinSVMvsGAM.m
This script runs the spin test to compare the map of summed absolute prediction weights from our machine learning model (Figure 3D) to a map of GAM effect size (Figure 4A)
-
Create hex spin plot with spintest/hexplots_20210301.R
This script creates a hex spin plot of GAM Loadings vs SVM weights (Figure 4B)
Part 5: Chromosomal enrichment analysis
Schaefer 1000, Fornito annotation strategy
-
Download data from https://figshare.com/articles/dataset/AHBAdata/6852911
Download
AHBAProcessed.zipandAHBAData.zip -
Read annotation file from figshare into Matlab
Annotation file is in
AHBAData/data/genes/parcellations/lh.Schaefer1000_7net.annot
[v, L, ct] = read_annotation(‘/cbica/projects/funcParcelSexDiff/inputData/genetics/sensitivity_analyses/parcellation/data/genes/parcellations/lh.Schaefer1000_7net.annot’)
save column 5 fromct.tableaslh_Schaefer1000_7net_cttable.csv
saveLaslh_Schaefer1000_7net_L.csv -
Save probe annotation file
Open
ROIxGene_Schaefer1000_INT.matin matlab
Gene = cell2table(probeInformation.GeneSymbol);
writetable(Gene, ‘/Users/sheilash/Desktop/projects/pfn_sex_diff/genetics/sensitivity_analyses/parcellation/data/genes/parcellations/schaefer1000/GeneSymbol.csv’)
-
Calculate chromosome enrichements and create Figure 5 with genetics/genetics/parcellation/schaefer1000_7networks/wSex_cor_gene_schaefer403_net7_20210224.R
This script parcellates map of SVM weights to schafer1000, merges gene and imaging data, removes missing parcels, calculates chromosomal enrichements (with significance testing), and creates figure 5
Schaefer 300, Fornito annotation strategy
-
Read annotation file from figshare into Matlab
Annotation file is in
AHBAData/data/genes/parcellations/lh.Schaefer300_7net.annot
[v, L, ct] = read_annotation(‘/cbica/projects/funcParcelSexDiff/inputData/genetics/sensitivity_analyses/parcellation/data/genes/parcellations/lh.Schaefer300_7net.annot’)
save column 5 fromct.tableaslh_Schaefer300_7net_cttable.csv
saveLaslh_Schaefer300_7net_L.csv -
Save probe annotation file
Open
ROIxGene_Schaefer300_INT.matin matlab
Gene = cell2table(probeInformation.GeneSymbol);
writetable(Gene, ‘/Users/sheilash/Desktop/projects/pfn_sex_diff/genetics/sensitivity_analyses/parcellation/data/genes/parcellations/schaefer300/GeneSymbol.csv’)
-
Calculate chromosome enrichements with genetics/genetics/sensitivity_analyses/fornito/wSex_cor_gene_schaefer300_net7_20201215.R
This script parcellates map of SVM weights to schafer300, merges gene and imaging data, removes missing parcels, and calculates chromosomal enrichements (with significance testing).
Schaefer 400, Seidlitz annotation strategy
-
Download all files from genetics/genetics/allen_processing/geneExpression_Repository
This repository contains scripts that align microarray gene expression data from AHBA donors to the left hemisphere of the Schaefer 400 parcellation. There were originally downloaded from geneExpression_Repository
-
Run genetics/genetics/allen_processing/geneExpression_Repository/main_sampleMatching.m
This script specifies the parcellation and mirroring strategy to be used. It then calls functions that download AHBA data, maps probes to genes, estimates gene expression from probes, estimates voxels location of each sample in T1 freesurfer space of each donor, and generates gene expression and coexpression matrices
Savegene_regional_expression_zscoredasgene_regional_expression_zscored.mat -
Calculate chromosome enrichements with genetics/genetics/sensitivity_analyses/wSexMultiTimes100_cor_gene_schaefer400_Seidlitz_20210308.R
This script parcellates map of SVM weights to schafer400 and calculates chromosomal enrichements (with significance testing).
Schaefer 400, Seidlitz annotation strategy, leave out female donor
-
Run genetics/genetics/allen_processing/geneExpression_Repository/main_sampleMatching_NoF.m
This script specifies the parcellation and mirroring strategy to be used. It then calls functions that download AHBA data, maps probes to genes, estimates gene expression from probes, estimates voxels location of each sample in T1 freesurfer space of each donor, and generates gene expression and coexpression matrices
Savegene_regional_expression_zscoredasgene_regional_expression_zscored_NoF.mat -
Calculate chromosome enrichements with genetics/genetics/sensitivity_analyses/wSexMultiTimes100_cor_gene_schaefer400_Seidlitz%20_NoF_20210308.R
This script parcellates map of SVM weights to schafer400 and calculates chromosomal enrichements (with significance testing).
Part 6: Gene Ontology and Cell Type Enrichement Analysis
Gene Ontology Analysis
-
This script parcellates map of SVM weights to schafer1000, merges gene and imaging data, removes missing parcels, and creates the ranked gene list
RankedGeneList_wSex100_Cor_schaefer1000Net7_20201123.csvfor GO enrichements,
-
Run gene ontology analysis with GOrilla
Note: the following analysis generating Supplementary Figure 2 was run on March 9, 2021
Settings should be as follows:
Step 1: homo sapiens
Step 2: Single ranked gene list
Step 3: upload RankedGeneList_wSex100_Cor_schaefer1000Net7_20201123.csv
Step 4: All
Advanced parameters:
> P-value threshold 10^-5
> SelectOutput results in Microsoft Excel format
> SelectShow output also in REVIGO
> UnselectInclude unresolved and duplicate genes in output
> UnselectRun GOrilla in fast mode
Cell Type Enrichement Analysis
-
This analysis uses data downloaded and prepared in Part 5, Schaefer1000, steps 1-3
-
Calculate cell type enrichements using PSP subtypes with genetics/genetics/cellTypes/schaefer1000_fornito/wSexMultiTimes100_cor_gene_cellTypeBrain_PSP_schaefer1000_fornito_20210102.R
This script parcellates map of SVM weights to schafer1000, merges gene and imaging data, removes missing parcels, and calculates cell type enrichements where cell-type were assigned according to categorizations determined by Seidlitz et al., 2020
-
Calculate cell type enrichements using Lake subtypes with genetics/genetics/cellTypes/schaefer1000_fornito/wSexMultiTimes100_cor_gene_cellTypeBrain_LakeAll_schaefer1000_fornito_20210515.R
This script parcellates map of SVM weights to schafer1000, merges gene and imaging data, removes missing parcels, calculates cell type enrichements where cell-type were assigned according to categorizations determined by Lake et al., 2018, and generates Supplementary Figure 2.
-
Determine which X-linked genes belong to cell type sub-classes with expression patterns that correlate with sex differences in topography genetics/genetics/parcellation/schaefer1000_7networks/sig_cellType_vs_Xlinked.R
This script merges chromosomal annotations and Lake cell-type assignments to determine which X-linked genes belong to the cell type sub-classes with expression patterns that correlate with sex differences in topography (as determined in step 3). Supplementary Table 1 was created using the information in
df_SigCelltype_X