Project Information and Reproducibility Guide
View the Project on GitHub PennLINC/thalamocortical_development
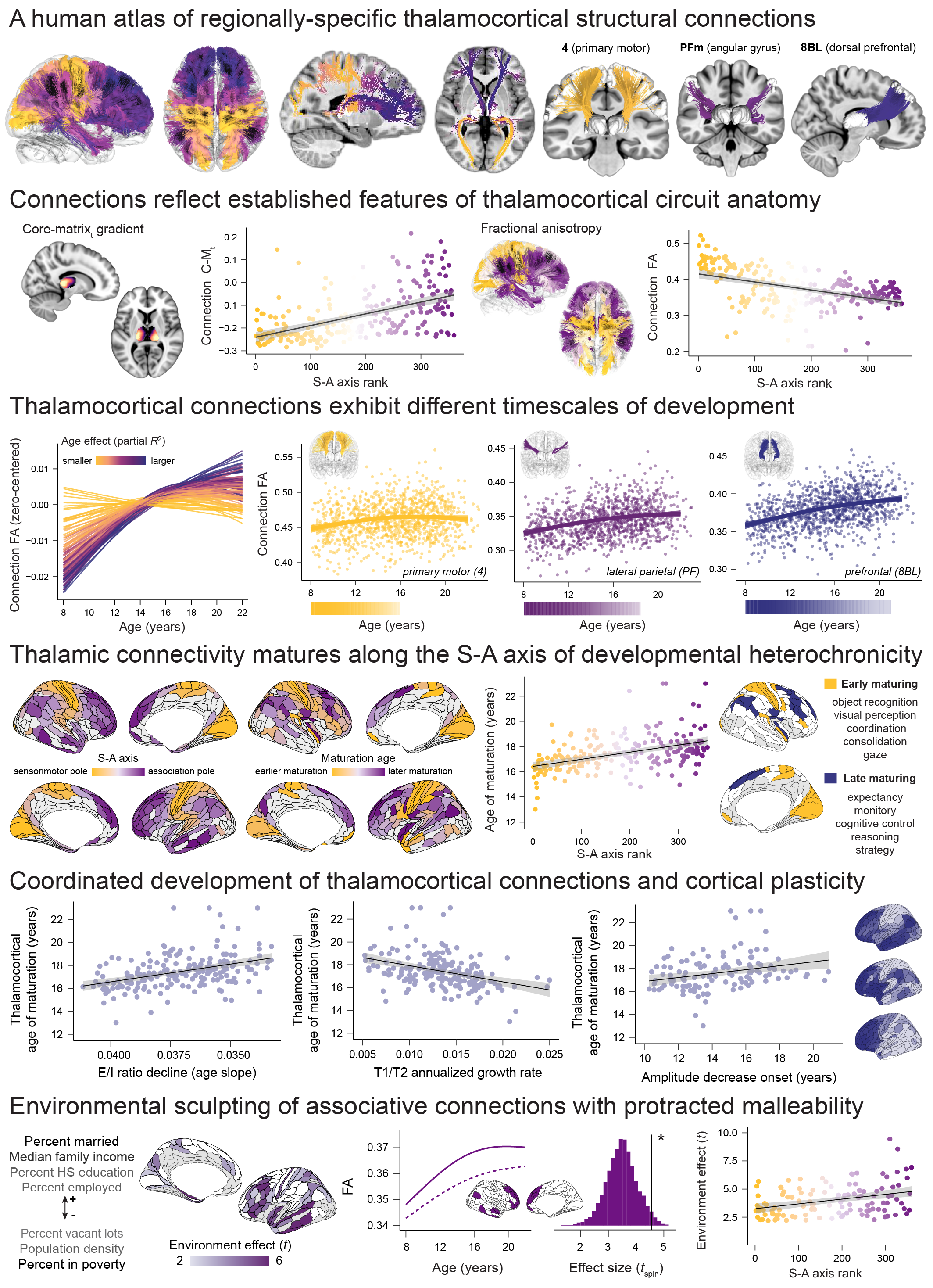
A Sensorimotor-Association Axis of Thalamocortical Connection Development
Project Lead
Valerie J. Sydnor
Faculty Lead
Theodore D. Satterthwaite
Analytic Replicators
Joelle Bagautdinova and Matthew Cieslak
Collaborators
Bart Larsen, Michael J. Arcaro, Deanna M. Barch, Dani S. Bassett, Aaron F. Alexander-Bloch, Philip A. Cook, Sydney Covitz, Alexandre R. Franco, Raquel E. Gur, Ruben C. Gur, Allyson P. Mackey, Kahini Mehta, Steven L. Meisler, Michael P. Milham, Tyler M. Moore, Eli J. Muller, David R. Roalf, Taylor Salo, Gabriel Schubiner, Jakob Seidlitz, Russell T. Shinohara, James M. Shine, Fang-Cheng Yeh
Project Start Date
December 2022
Current Project Status
Manuscript in revision
Datasets
PNC, HCPD, HBN
Github Repository
https://github.com/PennLINC/thalamocortical_development
Atlas of Human Thalamocortical Connections
The curated thalamocortical tractography atlas is available here! Detailed implementation instructions are provided for applying this tractography atlas to study thalamocortical connectivity in new data.
Cubic Project Directory
The project directory on cubic is: /cbica/projects/thalamocortical_development
The directory structure within the project directory is as follows:
code: directory with the thalamocortical_development github repo
cortical_anatomy: PNC and HCPD freesurfer tabulate anatomical statistics
figures: manuscript plots and images, compiled into final Figures
Maps: surface parcellation files (parcellations/), S-A axis github repo (S-A_ArchetypalAxis/), fluctuation amplitude development maps (boldamplitude_development/), myelin development maps (myelin_development/), E:I ratio development maps (EI_development/), thalamic Cpt gradient (thalamusgradient_CPt_muller/)
sample_info: sample demographics, environment data, and final project participant lists
software: project software
Templates: MNI template and HCP-1065 YA FIB templates
thalamocortical_results: GAM outputs for developmental and environmental effects
thalamocortical_structuralconnectivity/template: thalamocortical template tractography
thalamocortical_structuralconnectivity/individual: PNC, HCPD, and HBN autotrack outputs
qsirecon_0.16.0RC3: PNC and HCPD qsirecon clones with dsi-studio gqi and fib outputs
CODE DOCUMENTATION
The analytic and statistical workflow implemented in this research is described below; links to corresponding code on github are provided. This workflow begins with creation of an atlas of human thalamocortical connections. It continues with preprocessing and reconstruction of PNC, HCPD, and HBN diffusion MRI data, generation of individual-specific thalamocortical connections in youth datasets, and quantification and harmonization of thalamocortical connectivity metrics. It then transitions to fitting of generalized additive models to study relationships between thalamocortical connectivity, age, and the environment and describes analyses aimed at characterizing thalamocortical structural connectivity development and its influence on hierarchical cortical development along the sensorimotor-association axis.
Creation of an Atlas of Human Thalamocortical Connections (HCP-Young Adult)
A novel thalamocortical connectivity tractography atlas was generated using a high quality diffusion template derived from HCPYA data (N = 1,065, multi-shell acquisition parameters: b-values = 1000, 2000, 3000, 90 directions per shell, 1.25mm isotropic voxels). This population-average template, downloaded from here, is a 1.25 mm isotropic diffusion template in ICBM152 space generated with q-space diffeomorphic reconstruction (QSDR, the MNI-space version of generalized q-sampling imaging).
The tractography atlas was generated in the following steps:
- Generate thalamic tractography: Thalamic tractography was reconstructed with thalamocortical_structuralconnectivity/template/thalamic_tractography.sh by tracking 2 million streamlines with endpoints in the left thalamus and 2 million streamlines with endpoints in the right thalamus, based on the HCPYA diffusion template. Thalamic tractography was run with the following parameters:
--threshold_index=qa #use qa for thresholding fiber tracking --fa_threshold=0 #select a random qa value threshold as termination criterion for each streamline --turning_angle=0 #select a random turning angle threshold between 15-90 degrees as termination criterion for each streamline --step_size=0 #select a random step size from 0.5 to 1.5 voxels for each streamline --min_length=10 #minimum required length of streamlines --max_length=300 #maximum allowed length of streamlines --method=0 #streamline tracking --otsu_threshold=0.45 #Otsu's threshold --smoothing=1 #select a random smoothing amount between 0% to 95% for each streamline; smoothing uses previous propagation vector directional information --tip_iteration=0 #turn off topology-informed pruning --random_seed=1 #set the random seed for fiber tracking - Delineate regionally-specific thalamus-to-cortex connections: Structural connections between the thalamus and ipsilateral cortical regions were extracted from the thalamic tractography with thalamocortical_structuralconnectivity/template/thalamocortical_connections.sh using the HCP-MMP (glasser) atlas.
- Manually curate thalamocortical connections: All regionally-specific thalamocortical connections were visualized and manually edited, if needed, to ensure that atlas connections were compact, robust, comparable across hemispheres, and anatomically correct (based on available primate tract tracing or human diffusion MRI data). Manual editing included the removal of clear false positive streamlines and facilitated identification of thalamocortical connections that were very sparse or unreliable for removal from the atlas.
- Create skeletonized connections for autotracking: In order to facilitate use of the thalamocortical structural connectivity atlas with dsi-studio’s autotrack, each regionally-specific connection was skeletonized by deleting repeat streamlines with thalamocortical_structuralconnectivity/template/sparsify_connections.sh.
- Combine all regionally-specific thalamocortical connections into one tractography atlas: After generating finalized, regionally-specific structural connections between the thalamus and individual cortical areas, all connections were combined into one .tt.gz file for use in this study and for public distribution. The final version of the atlas only includes connections that could be robustly and reliably delineated in both the high-resolution HCPYA diffusion template and in individual participant’s data in PNC (single-shell, b=1000) and HCPD (multi-shell, bs=1500,3000). This atlas is provided in /results/thalamocortical_autotrack_template along with detailed implementation instructions.
Preprocessing and Reconstruction of Diffusion MRI Data (Philadelphia Neurodevelopment Cohort, HCP-Development, Healthy Brain Network)
Diffusion MRI data were preprocessed with qsiprep (0.14.2 for PNC and HBN; 0.16.1 for HCPD) as follows:
$ singularity run --cleanenv -B ${PWD} pennlinc-containers/.datalad/environments/qsiprep-${version}/image inputs/data prep participant --stop-on-first-crash --fs-license-file code/license.txt --skip-bids-validation --participant-label "$subid" --unringing-method mrdegibbs --output-resolution ${res} #res = 1.8 in PNC and HBN, 1.5 in HCPD
Diffusion MRI data were reconstructed using the dsi_studio_gqi reconstruction workflow with qsirecon (0.16.0RC3 for PNC, HCPD, HBN) as follows:
$ singularity run --cleanenv -B ${PWD} pennlinc-containers/.datalad/environments/qsiprep-0-16-0RC3/image inputs/data/qsiprep/qsiprep qsirecon participant --participant_label $subid -recon-input inputs/data/qsiprep/qsiprep --fs-license-file code/license.txt --stop-on-first-crash --recon-only --skip-odf-reports --freesurfer-input inputs/data/fmriprep/freesurfer --recon-spec ${PWD}/code/gqi_hsvs.json
Preprocessing and reconstruction workflows were executed with datalad using the template scripts in /qsiprep, including /PNC/qsiprep_call_PNC.sh, /HCPD/qsiprep_call_HCPD.sh,/HBN/qsiprep_call_HBN.sh,/PNC/qsirecon_call_PNC.sh, /HCPD/qsirecon_call_HCPD, and /HBN/qsirecon_call_HBN.sh. Datalad outputs were cloned for use in this project using the scripts in /datalad.
Delineation of Individual-Specific Thalamocortical Connections (Philadelphia Neurodevelopment Cohort, HCP-Development, Healthy Brain Network)
Person-specific thalamocortical structural connections were delineated for PNC, HCPD, and HBN participants using the thalamic tractography atlas and dsi-studio’s autotrack. A relatively stringent Hausdorff distance threhold was used; the selected threshold balanced the recovery of person-specific anatomy with mitigation of false positive streamlines and regionally non-specific streamlines. The script /thalamocortical_structuralconnectivity/individual/thalamocortical_autotrack.sh was run twice for every participant, first to generate participant -> template registration files (gqi.fib.gz.icbm152_adult.map.gz) for use with autotrack and then to reconstruct all thalamocortical connections.
For PNC, registration was accomplished with /thalamocortical_structuralconnectivity/individual/PNC/autotrack_registration_PNC.sh and autotrack tract generation was executed with /thalamocortical_structuralconnectivity/individual/PNC /run_autotrack_PNC.sh.
For HCPD, registration was accomplished with /thalamocortical_structuralconnectivity/individual/HCPD/autotrack_registration_HCPD.sh and autotrack tract generation was executed with /thalamocortical_structuralconnectivity/individual/HCPD/run_autotrack_HCPD.sh.
For HBN, registration was accomplished with /thalamocortical_structuralconnectivity/individual/HBN/autotrack_registration_HBN.sh and autotrack tract generation was executed with /thalamocortical_structuralconnectivity/individual/HBN/run_autotrack_HBN.sh.
- Autotrack was run with the following parameters:
--otsu_threshold=0.5 #Otsu's threshold for tracking --smoothing=1 #select a random smoothing amount between 0% to 95% for each streamline; smoothing uses previous propagation vector directional information --tolerance=10 #Hausdorff distance threhold --tip_iteration=0 #no topology-informed pruning --track_voxel_ratio=4 #the track-voxel ratio for the total number of streamline count. Increased from the default of 2 to facilitate better mapping (at the expense of greater computation time) --check_ending=0 #don't remove tracts that terminate in high anisotropy areas --export_stat=1 #write out connection statistics file --export_trk=1 #write out the reconstructed connection as a tractography file --yield_rate=0.0000001 #yield rate that must be met before fiber tracking is terminated and no output is generated --export_template_trk=1 #write out reconstructed connection in dsi-studio template spaceNote, thalamocortical_autotrack.sh was run twice (first to generate registration files, second to run autotrack on all atlas connections) due to how the threading parameter interacted with the registration process in dsi-studio. In recent versions of dsi-studio, greedy resource usage during registration was fixed, thus this procedure could now be run all in one step.
Quantification of Thalamocortical Connectivity Metrics
Diffusion MRI-derived connectivity metrics (FA, streamline count) and gene expression-derived thalamic core-matix gradient values were calculated in all thalamocortical connections in all participants, using the following code:
- Diffusion-derived connectivity metrics: Dataset-specific spreadsheets containing autotrack output statistics for every reconstructed thalamocortical connection were produced by running /tract_measures/diffusion_metrics/thalamocortical_dwimeasures.py with a “dataset” command line argument (i.e., “PNC”, “HCPD”, or “HBN”; example below). This python script takes in the dataset argument and generates a dataset-specific .csv file in long format with all diffusion metrics extracted for all tracts across all participants.
$ python thalamocortical_dwimeasures.py PNC
- Thalamus calbindin-parvalbumin CMt gradient values: A series of scripts were written in order to calculate each connection’s thalamic termination area CMt value. The CMt value represents a thalamocortical connection’s core-matrix gradient positioning (computed based on the relative expression of calbindin and parvalbumin genes). First, thalamocortical connection masks were registered to MNI space and CMt values were calculated in the connection’s thalamic termination area with /tract_measures/CPtgradient_values/thalamocortical_calculate_CPtvalues.sh; this script was called for every participant by executing /tract_measures/CPtgradient_values/thalamocortical_CPtvalues_jobs.sh with command line arguments of either “PNC” or “HCPD”. Then /tract_measures/CPtgradient_values/thalamocortical_CPtvalues.py was run with a “dataset” command line argument of either “PNC” or “HCPD”. This python script takes in a dataset command line argument and generates a dataset-specific .csv file in long format with CMt extracted for all tracts across all participants.
$ bash thalamocortical_CPtvalues_jobs.sh HCPD
$ python thalamocortical_CPtvalues.py HCPD
- Atlas overlap sensitivity and specificity values: Tractography reconstruction accuracy was assessed at the individual level for all thalamocortical connections by parameterizing the voxel-wise overlap between a participant’s thalamocortical connection and the same connection in the tractography atlas with measures of overlap sensitivity (true positive rate) and specificity (true negative rate). To compute these atlas overlap measure, template-space connection masks were first generated with /tract_measures/overlap/thalamocortical_templatetract_masks.sh, which was run on all participants by executing /tract_measures/overlap/thalamocortical_templatetract_masks_jobs.sh with a “dataset” command line argument, as above. Atlas overlap measures were then calculated with /tract_measures/overlap/thalamocortical_sensitivityspecificity.py which also takes in a command line dataset argument.
Sample Construction
1358 PNC participants, 640 (Lifespan 2.0 release) HCPD participants, and 1530 (Data Releases 1-9) HBN participants had dominant group diffusion MRI acquisitions (i.e., non-variant CuBIDS acquisitions) and were considered for inclusion in this research. The following exclusion criterion were then applied to generate the final samples of 1145 PNC participants, 572 HCPD participants, and 959 HBN participants:
- health history exclusions, for example history of cancer, MS, seizures, or incidentally-encountered brain structure abnormalities
- T1 quality exclusion, based on visual QC
- diffusion acquisition exclusion for missing runs (HCPD only)
- diffusion quality exclusion, based on the neighborhood correlation (nc) of the preprocessed, T1w-aligned diffusion data. Note, nc values differ by sampling scheme, thus different thresholds were used in PNC and HCPD. Thresholds were chosen based on nc histograms by selecting a value that cut off the low-quality (left-skewed) data tail.
- diffusion scan head motion exclusion, based on mean framewise displacement (threshold = 1 mm).
- An age exclusion (< 8 years old) was also applied to HCPD and HBN in order to match ages across samples and more appositely assess reproducibility of developmental results.
Final study samples were constructed following the criterion outlined above in /sample_construction/PNC/finalsample_PNC.Rmd, /sample_construction/HCPD/finalsample_HCPD.Rmd, and /sample_construction/HBN/finalsample_HBN.Rmd. This sample construction procedure utilized diffusion scan acqusition, quality, and head motion information provided in qsiprep ImageQC_dwi.csvs, which were collated into dataset-specific diffusion QC metric .csvs with /sample_construction/PNC/diffusion_qcmetrics_PNC.py, /sample_construction/HCPD/diffusion_qcmetrics_HCPD.py, and /sample_construction/HBN/diffusion_qcmetrics_HBN.py.
Generation of Dataset-Specific Analysis Dfs and Tract Lists
To facilitate analysis of participant-level thalamocortical connectivity data, dataset-specific demographics + diffusion dataframes and dataset-specific tract lists were generated.
- Dataset-specific demographics and diffusion dfs: Analytic dataframes were created for each developmental sample that include demographics and thalamocortical structural connectivity metrics for the final study sample. In HCPD and HBN, final sample thalamocortical metrics were harmonized with comBat to mitigate site effects. In all datasets, thalamocortical connections with < 5 streamlines were removed from final analytic dataframes at the participant-level; these connections were not included in further analyses. These steps (dataframe creation, harmonization, and streamline thresholding) were implemented with /sample_construction/PNC/tractmeasures_dfs_PNC.R, sample_construction/HCPD/tractmeasures_dfs_HCPD.R, and /sample_construction/HBN/tractmeasures_dfs_HBN.R.
- Dataset-specific tract lists: Only connections that could be reliably and robustly delineated at the individual level were studied in this work. To identify thalamocortical connections to study, tract_measures/tractlists/thalamocortical_tractlists.R and /tract_measures/tractlists/thalamocortical_tractlists_HBN.R were run. We implemented a connection-level streamline count threshold of >= 5 (as noted above) and a dataset-level connection inclusion threshold of >= 90% of participants.
Thalamocortical Connectivity: Coverage Characteristics and Circuit Motifs
Analyses were undertaken to examine tractography atlas cortical coverage and to anatomically validate identified connections. Anatomical validation included testing whether thalamocortical connectivity profiles reflected thalamic cellular classifications and hierarchical connectivity motifs. Analyses were conducted in /results/tract_anatomy/thalamocortical_connection_anatomy.Rmd; a knit version of thalamocortical_connection_anatomy.html can be viewed here.
- The total surface area and average sulcal depth of all glasser parcels was computed based on the fsaverage surface in /results/tract_anatomy/parcel_anatomy/glasser_parcel_anatomy.sh and /results/tract_anatomy/parcel_anatomy/glasser_parcel_anatomy.py.
Thalamocortical Connectivity: Hierarchical Development along the Sensorimotor-Association Axis
Analyses were undertaken in PNC and HCPD to characterize normative thalamocortical connection maturational patterns and to investigate whether thalamocortical connectivity development aligns with hierarchical axes of cortical developmental plasticity. Generalized additive models were used to delineate developmental trajectories, quantify age-related change, and identify the age of thalamocortical connectivity maturation. GAM analyses utilized the functions in /gam_functions/GAM_functions_thalamocortical.R, including:
- gam.fit.smooth: A function to fit a GAM (measure ~ s(smooth_var, k = knots, fx = set_fx) + covariates)) and save out statistics (F-value, partial R squared, p-value) and derivative-based characteristics (e.g., age window of significant increase, age of peak change)
- gam.smooth.predict: A function to predict fitted values of your dependent variable based on a GAM model and a prediction data frame using gratia::fitted_values
- gam.estimate.smooth: A function to estimate zero-averaged gam smooth estimates using gratia::smooth_estimates
- gam.derivatives: A function to compute derivatives for the smooth term from a main GAM model and for individual draws from the simulated posterior distribution; can return true model derivatives or posterior derivatives
- gam.factorsmooth.interaction: A function to fit a GAM with a factor-smooth interaction and obtain statistics for the interaction term
These functions were used to fit connection-wise developmental GAMs in /gam_functions/fit_ageGams.R. In general, this code applies the above functions to fit age GAMs for each thalamocortical connection of interest and saves out the results.
After fitting developmental models, the potential contribution of thalamocortical connectivity maturation to hierarchical development along the cortex’s S-A axis was investigated in /results/developmental_effects/thalamocortical_connectivity_development.Rmd, which can be viewed here. This investigation included: quantifying the similarity of developmental effects across datasets (PNC and HCPD), visualizing cortex-wide and region-specific thalamic connectivity developmental profiles and derivative heterochronicity, ascribing psychological functions to cortical regions with early and late maturing thalamic connections, testing how maturational timing varied across the S-A axis (and anatomical axes), and examining alignment between thalamocortical connection maturation and refinements in cortical E/I ratio, T1/T2 ratio, and BOLD fluctuation amplitude.
Thalamocortical Connectivity: Relationships with Neighborhood and Household Socioeconomic Conditions
GAMs were employed to explore associations between thalamocortical connection properties and youths’ household-level and neighborhood-level socioeconomic conditions (while controlling for developmental effects). Models were fit in /gam_functions/fit_envGams.R. Household-level SES was proxied by caregiver education and the income-to-needs ratio. Neighborhood-level socioeconomic advantage was determined based on a factor analysis of geocoded neighborhood environment indicators (e.g., median family income, percent employed, population density, percent in poverty). Environment GAMs used additional functions from /gam_functions/GAM_functions_thalamocortical.R, including:
- gam.fit.covariate: A function to fit a GAM (measure ~ s(smooth_var, k = knots, fx = set_fx) + covariates)) and save out statistics for the covariate of interest
- gam.smooth.predict.covariateinteraction: A function to predict fitted values of a measure for a given value of a covariate, using a varying coefficients smooth-by-linear covariate interaction
Significant associations between environmental conditions and thalamocortical connectivity were identified and spatial variability in their cortical embedding along S-A and anatomical axes was studied in /results/environment_effects/thalamocortical_connectivity_environment.Rmd, which can be viewed here.
Thalamocortical Connectivity: Replication of Developmental and Environmental Effects in a Clinical Sample of Youth with Psychopathology
After obtaining the core set of findings in PNC and HCPD, we evaluated the generalizability of results to the HBN, a sample of youth with clinically-significant youth psychopathology. Developmental GAMs for the HBN were fit in /gam_functions/fit_ageGams_HBN.R and environmental gams were run in /gam_functions/fit_envGams_HBN.R.
Code investigating alignment of developmental and environmental effects to the S-A axis in the HBN is in /results/HBN_replication/thalamocortical_connectivity_HBNreplication.Rmd, which can be viewed as an html here!
Developmental Sensitivity Analyses
Three sensitivity analyses were conducted to ensure that developmental findings were not being driven by potential anatomical and methodological confounds. Sensitivity analyses considered the potential impact of cortical region endpoint anatomy indexed by regional surface area, diffusion data quality measured by tSNR, and tractography reconstruction accuracy determined by atlas overlap sensitivity (true positive rate).
- For the surface area analyses, T1w images were analyzed with FreeSurfer 6.0.1. freesurfer_tabulate was then applied to processed anatomical data to calculate the surface area of regions included in the HCP-MMP atlas.
- For the tSNR analysis, the voxel-wise map of b=0 shell tSNR was obtained from FSL eddy outputs generated during QSIPrep. The average tSNR was then computed in masks of all participants’ thalamocortical connections by running /tract_measures/snr/thalamocortical_SNR_jobs.sh, which launches /tract_measures/snr/thalamocortical_calculate_SNR.sh for each participant. SNR data were collated into dataset-specific csvs using /tract_measures/snr/thalamocortical_SNRvalues.py.
- For the atlas overlap analysis, sensitivity (true positive rates) were used.
To create dataframes with connection-specific sensitivity variables while excluding connections with < 5 streamlines from analysis, /sample_construction/PNC/tractmeasures_sensitivity_PNC.R was run along with /sample_construction/HCPD/tractmeasures_sensitivity_HCPD.R. Sensitivity development GAMs were then fit using these dataframes in /gam_functions/fit_sensitivityGams.R. Finally, the results of sensitivity analyses are presented in /results/sensitivity_analyses/thalamocortical_development_sensitivity.Rmd which can be viewed here.
DIFFUSION DATA AVAILABILITY
This study used existing developmental data from the Philadelphia Neurodevelopmental Cohort, the Lifespan 2.0 Human Connectome Project-Development release, and data releases 1-9 from the Healthy Brain Network. Raw diffusion data from these studies are accessible from the links provided below:
- PNC: https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000607.v3.p2
- HCPD: https://nda.nih.gov/ccf
- HBN: https://fcon_1000.projects.nitrc.org/indi/cmi_healthy_brain_network/
PennLINC is currently in the process of gaining all permissions necessary to make preprocessed diffusion data and derivatives from the PNC, HCPD, and HBN datasets publicly available. This section will be updated when data are publicly released! In the meantime, you can access processed structural and functional MRI data from these three datasets at the Reproducible Brain Charts website.
PROJECT SOFTWARE
The following external software was used in this project:
- qsiprep versions 0.14.2 and 0.16.1 https://hub.docker.com/r/pennbbl/qsiprep
- dsistudio container version chen-2023-02-27, installed via singularity with /software_installation/build_dsistudio_image.sh and accessible here
- datalad version 0.18.1, installed via anaconda
- connectome workbench v.1.5.0, downloaded from humanconnectome.org into /software/workbench
- rotate_parcellation algorithm for parcel-based spin testing, cloned from the rotate_parcellation github into /software/rotate_parcellation
- freesurfer version 6.0.0 (freesurfer-Darwin-OSX-stable-pub-v6.0.0-2beb96c), downloadable from freesurfer’s older releases archives
- R version 4.2.3; packages: dplyr, plyr, tidyr, tidyverse, purrr, tibble, mgcv, gratia, ggplot2, ggseg, ggsegGlasser, ggnewscale, scales, cifti, PupillometryR, car, rstatix, Hmisc, matrixStats, cocor, reshape2, EnvStats, neuroCombat, datawizard
Analyses were conducted on the CUBIC cluster at the University of Pennsylvania, a RedHat Enterprise Linux-based HPC cluster.
And Fin :)