Diffusion Data Preprocessing
Citation
Cieslak, Matthew, Philip A Cook, Xiaosong He, Fang-Cheng Yeh, Thijs Dhollander, Azeez Adebimpe, Geoffrey K Aguirre, et al. 2021. “QSIPrep: An Integrative Platform for Preprocessing and Reconstructing Diffusion Mri Data.” Nature Methods 18 (7). Nature Publishing Group US New York: 775–78. https://doi.org/10.1038/s41592-021-01185-5.
Cieslak, Matthew, Philip A Cook, Xiaosong He, Fang-Cheng Yeh, Thijs Dhollander, Azeez Adebimpe, Geoffrey K Aguirre, et al. 2021. “QSIPrep: An Integrative Platform for Preprocessing and Reconstructing Diffusion Mri Data.” Nature Methods 18 (7). Nature Publishing Group US New York: 775–78. https://doi.org/10.1038/s41592-021-01185-5.
More citations (Click to expand/minimize)
Abraham, Alexandre, Fabian Pedregosa, Michael Eickenberg, Philippe Gervais, Andreas Mueller, Jean Kossaifi, Alexandre Gramfort, Bertrand Thirion, and Gael Varoquaux. 2014. “Machine Learning for Neuroimaging with Scikit-Learn.” Frontiers in Neuroinformatics 8. https://doi.org/10.3389/fninf.2014.00014.
Andersson, Jesper LR, Mark S Graham, Enikő Zsoldos, and Stamatios N Sotiropoulos. 2016. “Incorporating Outlier Detection and Replacement into a Non-Parametric Framework for Movement and Distortion Correction of Diffusion Mr Images.” Neuroimage 141. Elsevier: 556–72.
Andersson, Jesper LR, and Stamatios N Sotiropoulos. 2016. “An Integrated Approach to Correction for Off-Resonance Effects and Subject Movement in Diffusion Mr Imaging.” Neuroimage 125. Elsevier: 1063–78.
Billot, Benjamin, Douglas N Greve, Oula Puonti, Axel Thielscher, Koen Van Leemput, Bruce Fischl, Adrian V Dalca, Juan Eugenio Iglesias, and others. 2023. “SynthSeg: Segmentation of Brain Mri Scans of Any Contrast and Resolution Without Retraining.” Medical Image Analysis 86. Elsevier: 102789.
Billot, Benjamin, Colin Magdamo, You Cheng, Steven E Arnold, Sudeshna Das, and Juan Eugenio Iglesias. 2023. “Robust Machine Learning Segmentation for Large-Scale Analysis of Heterogeneous Clinical Brain Mri Datasets.” Proceedings of the National Academy of Sciences 120 (9). National Acad Sciences: e2216399120.
Cordero-Grande, Lucilio, Daan Christiaens, Jana Hutter, Anthony N Price, and Jo V Hajnal. 2019. “Complex Diffusion-Weighted Image Estimation via Matrix Recovery Under General Noise Models.” Neuroimage 200. Elsevier: 391–404. https://doi.org/10.1016/j.neuroimage.2019.06.039.
Garyfallidis, Eleftherios, Matthew Brett, Bagrat Amirbekian, Ariel Rokem, Stefan Van Der Walt, Maxime Descoteaux, and Ian Nimmo-Smith. 2014. “Dipy, a Library for the Analysis of Diffusion Mri Data.” Frontiers in Neuroinformatics 8. Frontiers: 8.
Gorgolewski, K., C. D. Burns, C. Madison, D. Clark, Y. O. Halchenko, M. L. Waskom, and S. Ghosh. 2011. “Nipype: A Flexible, Lightweight and Extensible Neuroimaging Data Processing Framework in Python.” Frontiers in Neuroinformatics 5: 13. https://doi.org/10.3389/fninf.2011.00013.
Gorgolewski, Krzysztof J., Oscar Esteban, Christopher J. Markiewicz, Erik Ziegler, David Gage Ellis, Michael Philipp Notter, Dorota Jarecka, et al. 2018. “Nipype.” Software. Zenodo. https://doi.org/10.5281/zenodo.596855.
Hoopes, Andrew, Jocelyn S Mora, Adrian V Dalca, Bruce Fischl, and Malte Hoffmann. 2022. “SynthStrip: Skull-Stripping for Any Brain Image.” NeuroImage 260. Elsevier: 119474.
Lee, Hong-Hsi, Dmitry S Novikov, and Els Fieremans. 2021. “Removal of Partial Fourier-Induced Gibbs (Rpg) Ringing Artifacts in Mri.” Magnetic Resonance in Medicine 86 (5). Wiley Online Library: 2733–50.
Power, Jonathan D., Anish Mitra, Timothy O. Laumann, Abraham Z. Snyder, Bradley L. Schlaggar, and Steven E. Petersen. 2014. “Methods to Detect, Characterize, and Remove Motion Artifact in Resting State fMRI.” NeuroImage 84 (Supplement C): 320–41. https://doi.org/10.1016/j.neuroimage.2013.08.048.
Tournier, J-Donald, Robert Smith, David Raffelt, Rami Tabbara, Thijs Dhollander, Maximilian Pietsch, Daan Christiaens, Ben Jeurissen, Chun-Hung Yeh, and Alan Connelly. 2019. “MRtrix3: A Fast, Flexible and Open Software Framework for Medical Image Processing and Visualisation.” Neuroimage 202. Elsevier: 116137. https://doi.org/10.1016/j.neuroimage.2019.116137.
Tustison, N. J., B. B. Avants, P. A. Cook, Y. Zheng, A. Egan, P. A. Yushkevich, and J. C. Gee. 2010. “N4ITK: Improved N3 Bias Correction.” IEEE Transactions on Medical Imaging 29 (6): 1310–20. https://doi.org/10.1109/TMI.2010.2046908.
Veraart, Jelle, Els Fieremans, and Dmitry S Novikov. 2016. “Diffusion Mri Noise Mapping Using Random Matrix Theory.” Magnetic Resonance in Medicine 76 (5). Wiley Online Library: 1582–93.
Veraart, Jelle, Dmitry S Novikov, Daan Christiaens, Benjamin Ades-Aron, Jan Sijbers, and Els Fieremans. 2016. “Denoising of Diffusion Mri Using Random Matrix Theory.” NeuroImage 142. Elsevier: 394–406.
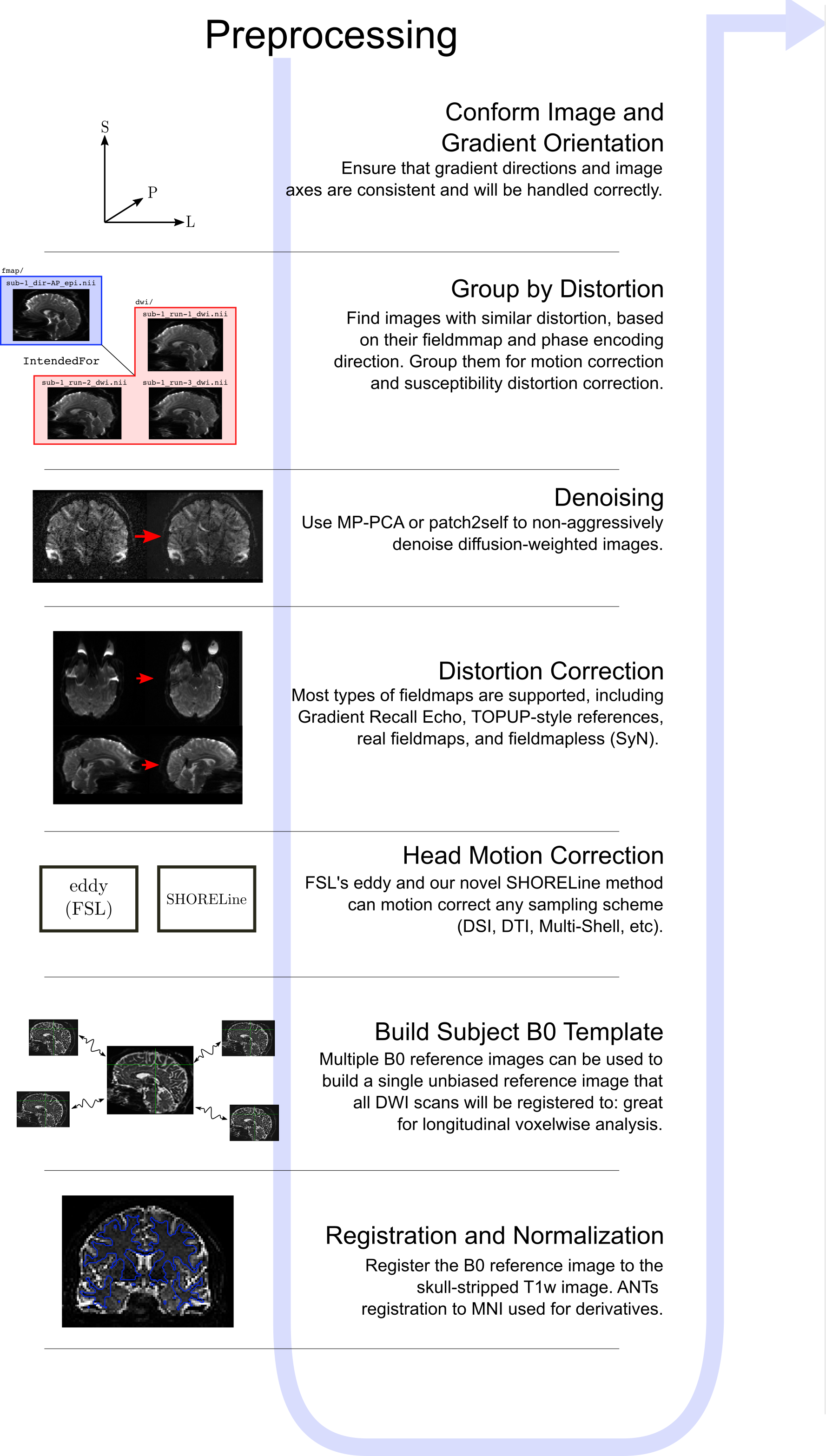
Diffusion data were preprocessed using QSIPrep.
Diffusion data were denoised using the Marchenko-Pastur PCA (MP-PCA) method, followed by Gibbs ringing removal, and correction for field inhomogeneity (N4). Motion and Eddy current distortions were corrected with FSL’s eddy. Final DWI data were resampled to ACPC space.
IMPORTANT: The exact QSIPrep pipeline is documented in the boilerplate located at
The exact
<QSIPrep_output>/logs/CITATION.html. If the data were processed with BABS (i.e., were zipped), refer to Get Data for details on unzipping <QSIPrep_output> to access the boilerplate file.
The exact
singularity run or apptainer run command can be found in the report.html file. We typically use the same CLI arguments across studies. If a study uses different arguments, they will be listed in the Command tab of the pipeline on the study-specific page.
Quality Control
QSIPrep produces *desc-image_qc.tsv files for QC dMRI data, more details here.